Abstract
Many organs with a high cell turnover (for example, skin, intestine and blood) are composed of short-lived cells that require continuous replenishment by somatic stem cells1,2. Ageing results in the inability of these tissues to maintain homeostasis and it is believed that somatic stem-cell ageing is one underlying cause of tissue attrition with age or age-related diseases. Ageing of haematopoietic stem cells (HSCs) is associated with impaired haematopoiesis in the elderly3,4,5,6. Despite a large amount of data describing the decline of HSC function on ageing, the molecular mechanisms of this process remain largely unknown, which precludes rational approaches to attenuate stem-cell ageing. Here we report an unexpected shift from canonical to non-canonical Wnt signalling in mice due to elevated expression of Wnt5a in aged HSCs, which causes stem-cell ageing. Wnt5a treatment of young HSCs induces ageing-associated stem-cell apolarity, reduction of regenerative capacity and an ageing-like myeloid–lymphoid differentiation skewing via activation of the small Rho GTPase Cdc42. Conversely, Wnt5a haploinsufficiency attenuates HSC ageing, whereas stem-cell-intrinsic reduction of Wnt5a expression results in functionally rejuvenated aged HSCs. Our data demonstrate a critical role for stem-cell-intrinsic non-canonical Wnt5a signalling in HSC ageing.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Morrison, S. J., Uchida, N. & Weissman, I. L. The biology of hematopoietic stem cells. Annu. Rev. Cell Dev. Biol. 11, 35–71 (1995)
Fuchs, E. & Segre, J. A. Stem cells: a new lease on life. Cell 100, 143–155 (2000)
Rossi, D. J. et al. Cell intrinsic alterations underlie hematopoietic stem cell aging. Proc. Natl Acad. Sci. USA 102, 9194–9199 (2005)
Rossi, D. J., Jamieson, C. H. & Weissman, I. L. Stems cells and the pathways to aging and cancer. Cell 132, 681–696 (2008)
Geiger, H. & Van Zant, G. The aging of lympho-hematopoietic stem cells. Nature Immunol. 3, 329–333 (2002)
Geiger, H. & Rudolph, K. L. Aging in the lympho-hematopoietic stem cell compartment. Trends Immunol. 30, 360–365 (2009)
Conboy, I. M., Conboy, M. J., Smythe, G. M. & Rando, T. A. Notch-mediated restoration of regenerative potential to aged muscle. Science 302, 1575–1577 (2003)
Conboy, I. M. et al. Rejuvenation of aged progenitor cells by exposure to a young systemic environment. Nature 433, 760–764 (2005)
Brack, A. S. et al. Increased Wnt signaling during aging alters muscle stem cell fate and increases fibrosis. Science 317, 807–810 (2007)
Liu, H. et al. Augmented Wnt signaling in a mammalian model of accelerated aging. Science 317, 803–806 (2007)
Malhotra, S. & Kincade, P. W. Wnt-related molecules and signaling pathway equilibrium in hematopoiesis. Cell Stem Cell 4, 27–36 (2009)
Nemeth, M. J., Topol, L., Anderson, S. M., Yang, Y. & Bodine, D. M. Wnt5a inhibits canonical Wnt signaling in hematopoietic stem cells and enhances repopulation. Proc. Natl Acad. Sci. USA 104, 15436–15441 (2007)
Duncan, A. W. et al. Integration of Notch and Wnt signaling in hematopoietic stem cell maintenance. Nature Immunol. 6, 314–322 (2005)
Luis, T. C., Naber, B. A., Fibbe, W. E., van Dongen, J. J. & Staal, F. J. Wnt3a nonredundantly controls hematopoietic stem cell function and its deficiency results in complete absence of canonical Wnt signaling. Blood 116, 496–497 (2010)
Luis, T. C. et al. Canonical Wnt signaling regulates hematopoiesis in a dosage-dependent fashion. Cell Stem Cell 9, 345–356 (2011)
Staal, F. J. & Luis, T. C. Wnt signaling in hematopoiesis: crucial factors for self-renewal, proliferation, and cell fate decisions. J. Cell. Biochem. 109, 844–849 (2010)
Schlessinger, K., Hall, A. & Tolwinski, N. Wnt signaling pathways meet Rho GTPases. Genes Dev. 23, 265–277 (2009)
Schlessinger, K., McManus, E. J. & Hall, A. Cdc42 and noncanonical Wnt signal transduction pathways cooperate to promote cell polarity. J. Cell Biol. 178, 355–361 (2007)
Sugimura, R. et al. Noncanonical Wnt signaling maintains hematopoietic stem cells in the niche. Cell 150, 351–365 (2012)
Florian, M. C. et al. Cdc42 activity regulates hematopoietic stem cell aging and rejuvenation. Cell Stem Cell 10, 520–530 (2012)
Geiger, H., de Haan, G. & Florian, M. C. The ageing haematopoietic stem cell compartment. Nature Rev. Immunol. 13, 376–389 (2013)
Peterson, J. R., Lebensohn, A. M., Pelish, H. E. & Kirschner, M. W. Biochemical suppression of small-molecule inhibitors: a strategy to identify inhibitor targets and signaling pathway components. Chem. Biol. 13, 443–452 (2006)
Liang, H. et al. Wnt5a inhibits B cell proliferation and functions as a tumor suppressor in hematopoietic tissue. Cancer Cell 4, 349–360 (2003)
Kuhl, M., Sheldahl, L. C., Park, M., Miller, J. R. & Moon, R. T. The Wnt/Ca2+ pathway: a new vertebrate Wnt signaling pathway takes shape. Trends Genet. 16, 279–283 (2000)
Zou, P. et al. p57Kip2 and p27Kip1 cooperate to maintain hematopoietic stem cell quiescence through interactions with Hsc70. Cell Stem Cell 9, 247–261 (2011)
Li, J. et al. WNT5A antagonizes WNT/β-catenin signaling and is frequently silenced by promoter CpG methylation in esophageal squamous cell carcinoma. Cancer Biol. Ther. 10, 617–624 (2010)
Kohler, A. et al. Altered cellular dynamics and endosteal location of aged early hematopoietic progenitor cells revealed by time-lapse intravital imaging in long bones. Blood 114, 290–298 (2009)
Adolfsson, J. et al. Identification of Flt3+ lympho-myeloid stem cells lacking erythro-megakaryocytic potential a revised road map for adult blood lineage commitment. Cell 121, 295–306 (2005)
Yang, L. et al. Cdc42 critically regulates the balance between myelopoiesis and erythropoiesis. Blood 110, 3853–3861 (2007)
Daria, D. et al. The retinoblastoma tumor suppressor is a critical intrinsic regulator for hematopoietic stem and progenitor cells under stress. Blood 111, 1894–1902 (2008)
Li, Z. et al. Predictable and efficient retroviral gene transfer into murine bone marrow repopulating cells using a defined vector dose. Exp. Hematol. 31, 1206–1214 (2003)
Acknowledgements
We thank G. Van Zant and J. A. Cancelas for advice and critical reading of the manuscript. We thank F. Kirchhoff and D. van der Merwe for cell sorting support, A. Rück and the Institut für Lasertechnologien in der Medizin und Meßtechnik of Ulm University for support with confocal microscopy, and the Mouse and Cancer Core in Cincinnati and the Tierforschungszentrum of the University of Ulm for supporting our animal work. The work in the laboratory of H.G. is supported by grants from the Deutsche Forschungsgemeinschaft KFO 142, GE2063/1 and SFB 1074, the German Federal Ministry of Education and Research within its joint research project SyStaR (also to H.A.K. and K.S.-K.), the Excellence program of the Baden-Württemberg Foundation, the National Institute of Health, HL076604, DK077762 and AG040118, the Edward P. Evans foundation and the European Commission (FP7 Marie Curie Initial Training Network MARRIAGE). M.C.F. is supported by a ‘Bausteinprogramm’ of the Department of Medicine of Ulm University.
Author information
Authors and Affiliations
Contributions
M.C.F. and H.G. designed and interpreted experiments and wrote the manuscript. M.C.F., K.J.N., B.Ü., C.E., M.S. I.A. and R.A.J.O. performed and analysed experiments. H.G. helped to perform transplantation experiments. V.V. helped with the Wnt5a mouse colony and performed inverse transplantation experiments and prepared sorted stroma CD45− cells. K.D. and G.M. assisted in transplantation procedures and supported in cell sorting and flow analysis procedures. H.A.K. performed expression and database analyses. Y.Z. and K.S.-K. assisted in designing and interpreting experiments.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
Extended Data Figure 1 Increased expression of Wnt5a in aged LT-HSCs results in a shift from canonical to non-canonical Wnt signalling.
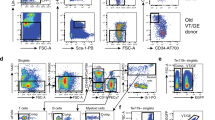
a, Reverse transcriptase real-time PCR analysis of Wnt5a transcript levels in young (10-week-old), middle-aged (10-month-old) and aged (24-month-old) LT-HSCs (Lin−c-kit+Sca-1+Flk2−CD34− bone marrow cells) sorted from C57BL/6 mice. Data are expressed as fold increased compared to the lowest expressed transcript arbitrarily set to 1. Wnt5a mRNA is barely detectable in young LT-HSCs and is markedly upregulated in middle-aged and aged LT-HSCs. Data were analysed with the 2−ΔΔCt method and plotted on a logarithmic scale. Bars are mean + 1 s.e.; n = 3, *P < 0.05. b, Reverse transcriptase real-time PCR analysis of Wnt5a transcript levels in young (10-week-old), middle-aged (10-month-old) and aged (24-month-old) Lin− bone marrow cells from C57BL/6 mice. Data are expressed as fold increased compared to the lowest expressed transcript arbitrarily set to 1. Wnt5a mRNA is barely detectable in young Lin− cells and is upregulated in middle-aged and aged LT-HSCs. Data were analysed with the 2−ΔΔCt method and plotted on a logarithmic scale. Scale bars represent results of one set of samples. The experiment was repeated twice with similar results. c, Reverse transcriptase real-time PCR analysis of Wnt5a transcript levels in young (10-week-old) and aged (24-month-old) LT-HSCs (Lin−c-Kit+Sca-1+Flk2−CD34− bone marrow cells) sorted from C57BL/6 and DBA/2 mice. Data are expressed as fold increased compared to the lowest expressed transcript arbitrarily set to 1. Data were analysed with the 2−ΔΔCt method and plotted on a logarithmic scale. Error bars are mean + 1 s.e.; n = 3, *P < 0.05, **P < 0.01. d, Representative three-dimensional confocal picture of Wnt5a distribution in an aged LT-HSC. The nucleus is stained with DAPI. Three-dimensional localization of Wnt5a was analysed by scanning the cells along the z-axis and acquiring a picture of the xy-plane every 0.7 μm. Three-dimensional images were then reconstructed by using Volocity v6.0 software. e, Representative immunofluorescence picture of Wnt5a (green) membrane distribution (non-permeabilized cells) in young and aged LT-HSCs. Immunofluorescence pictures are shown as overlap with the phase contrast image. Scale bar, 5 μm. f, Representative immunofluorescence picture of Wnt5a (green) and clathrin (red) localization in young and aged LT-HSCs. Pictures are shown on a dark background and as overlap with DAPI (staining nuclei). Scale bar, 5 μm. g, Representative expression of Wnt5a in MEFs (mouse embryonic fibroblasts) and aged LT-HSCs from Wnt5a+/+ mice determined by immunofluorescence. Wnt5a fluorescence signal is not detected when MEFs from Wnt5a−/−mice are stained with the same procedure. Wnt5a pictures are shown on a dark background and as overlap with DAPI (blue, staining nuclei) and phase contrast images. Scale bar, 10 μm. h, Representative FACS dot plots of LT-HSCs (Lin−c-Kit+Sca-1+Flk2−CD34−), ST-HSCs (Lin−c-Kit+Sca-1+Flk2−CD34+), LMPPs (Lin−c-Kit+Sca-1+Flk2+CD34+), LSKs (Lin−c-Kit+Sca-1+) and LKs (Lin−c-Kit+Sca-1−) gating strategy of young and aged lineage-depleted bone marrow cells. i, Representative FACS histograms of β-catenin expression in young, aged and young Wnt5a-treated LT-HSCs, ST-HSCs, LMPPs and LSKs.
Extended Data Figure 2 Wnt5a activates Cdc42 inducing ageing-like phenotypes in young LT-HSCs.
a, Representative distribution of Cdc42, tubulin and Per2 (staining the centrosome) in young control, young Wnt5a-treated (100 ng ml−1) and young Wnt5a (100 ng ml−1) + casin (5 µM)-treated LT-HSCs determined by immunofluorescence. Scale bar, 5 μm. Shown are also representative fluorescence intensity plots obtained by collecting pixel intensity through the section of the cell as indicated by the dotted line in the corresponding merge picture. b, Representative distribution of Cdc42, tubulin and NCam2 (membrane protein) in young control, young Wnt5a-treated (100 ng ml−1) and aged LT-HSCs determined by immunofluorescence. Scale bar, 5 μm. c, Graph of the percentage of young control, young Wnt5a-treated (100 ng ml−1) and aged LT-HSCs with a polar distribution of NCam2. Shown are mean + 1 s.e., n = 4; ∼200–300 LT-HSCs scored per sample in total. *P < 0.001. d, Reverse transcriptase real-time PCR analysis of Cdc42, Rhou, Rac1, Rac2, Rhoa, Rhoj, Rhov and Rhoq transcript levels in young, aged and young Wnt5a-treated (100 ng ml−1, 16 h treatment) LT-HSCs. Rhou and Rhov transcripts were below detection limits in all the assayed samples (ND). Data are expressed as fold difference compared to the expression of Cdc42 mRNA in young LT-HSCs arbitrarily set to 1. Data were analysed with the 2−ΔΔCt method and plotted on a linear scale. Bars are mean + 1 s.e.; n = 3, *P < 0.05. e, Schematic representation of the experimental set-up for the transplantation. Recipient mice were analysed 24 weeks after transplant.
Extended Data Figure 3 Wnt5a haploinsufficient mice present with attenuated HSC ageing.
a–d, White blood (WB) cell count (a), red blood (RB) cell count (b), haemoglobin (Hb) dosage (c) and lymphocyte cell count (d) in peripheral blood of young and aged Wnt5a+/− and Wnt5a+/+ mice. *P < 0.05; shown are mean + 1 s.e., n = 5. e, Percentage of LSKs among Lin− cells in bone marrow of young and aged Wnt5a+/− and Wnt5a+/+ mice. *P < 0.05; shown are mean + 1 s.e., n = 5. f, Reverse transcriptase real-time PCR analysis of Wnt5a transcript levels in young and aged LT-HSCs and young and aged collagenase-digested and sorted CD45− cells (stroma cells). Data are expressed as fold difference compared to the expression in young LT-HSCs arbitrarily set to 1. Wnt5a mRNA shows significantly increased expression in stroma CD45− cells when compared to young and aged LT-HSCs. In contrast to the situation in LT-HSCs, young stroma CD45− cells express higher levels of Wnt5a mRNA than aged stroma CD45− cells. Data were analysed with the 2−ΔΔCt method and plotted on a logarithmic scale. Error bars are mean + 1 s.e.; n = 4, *P < 0.05. g, Schematic representation of the experimental set-up for transplantation. Young donor (Ly5.1+) bone marrow cells were transplanted into recipient (Ly5.2+) young Wnt5a+/− and Wnt5a+/+ mice. Recipient mice were killed and analysed 20 months after transplant. h, i, Percentage of engrafted cells, B220+, CD3+ and myeloid cells among donor-derived Ly5.1+ cells in peripheral blood (h) and bone marrow (i) 20 months after transplants. Columns are mean values + 1 s.e., n = 5. j, Percentage of donor-derived LSKs among donor-derived Lin− cells in bone marrow of Wnt5a+/− and Wnt5a+/+ recipient mice 20 months after transplant. Columns show mean + 1 s.e.m., n = 5. k, Percentage of donor-derived LT-HSCs, ST-HSCs and LMPPs among donor-derived LSKs in Wnt5a+/− and Wnt5a+/+ recipient mice 20 months after transplant. Columns are mean values + 1 s.e. n = 5. l, m, Percentage of LT-HSCs polarized for Cdc42 (l) and tubulin (m) in young and aged Wnt5a+/+ and Wnt5a+/− mice. Shown are mean + 1 s.e., n = 4 and 200 cells scored per sample in total. *P < 0.01 versus young Wnt5a+/+ and P < 0.05 versus young Wnt5a+/− and aged Wnt5a+/− mice.
Extended Data Figure 4 Validation of the knockdown efficiency in 3T3 fibroblast cells and aged and young Lin− bone marrow cells.
a, Transduced fibroblast cells were sorted and analysed by western blot for Wnt5a protein levels. Wnt5a protein levels were normalized on actin. Three different Wnt5a knockdown vectors (3a-GFP+, 4a-GFP+, 5b-GFP+) were tested and Wnt5a protein levels were compared to non-targeting transduced fibroblasts (NT-GFP+) and to untransduced cells sorted as GFP− from the initial mixed culture (3a-GFP−, 4a-GFP−, 5b-GFP−). b, Transduced fibroblast cells were sorted and analysed by reverse transcriptase real-time PCR for Wnt5a mRNA levels. Wnt5a mRNA levels are normalized to actin mRNA levels. Three different Wnt5a knockdown vectors (3a-GFP+, 4a-GFP+, 5b-GFP+) were tested and Wnt5a transcript levels were compared to non-targeting transduced fibroblasts (NT-GFP+). c, Not pre-stimulated transduced Wnt5aKD or non-targeted aged Lin−bone marrow cells were sorted and analysed by reverse transcriptase real-time PCR for Wnt5a mRNA levels. Wnt5a mRNA levels are normalized on Gapdh mRNA levels. d, Percentage of B220+, CD3+ and myeloid cells among donor-derived cells in peripheral blood 24 weeks after transplant. *P < 0.05; shown are mean values + 1 s.e. Mice were considered as engrafted when the percentage of Ly5.2+ cells in peripheral blood was higher than 1.0 and contribution was detected for all peripheral blood lineages. Data are based on two different lentiviral infection/transplant experiments with 5–7 recipient mice per group (for example, n = 10). e, Schematic representation of the experimental set-up for transplantation of Wnt5a knock down (Wnt5aKD), Wnt5a non-targeting (Wnt5a-NT) and untransduced young haematopoietic progenitor/stem cells. Young donor (Ly5.2+) lineage-negative (Lin−) bone marrow cells were infected with the indicated lentiviral vectors or left untransduced. Infected cells were sorted based on GFP expression. Cells (1–3 × 105 Ly5.2+) were transplanted into recipients (Ly5.1+). Recipient mice were analysed 12–16 weeks after transplant. f, Percentage of engrafted donor-derived cells in peripheral blood 12–16 weeks after transplant. Shown are mean values + 1 s.e. Mice were considered as engrafted when the percentage of Ly5.2+ cells in peripheral blood was higher than 1.0 and contribution was detected for all peripheral blood lineages. Data are based on two different lentiviral infection/transplant experiments with 3 recipient mice per group (for example, n = 3 for Wnt5aKD and Wnt5a-NT mice and n = 6 for untransduced mice). g, Percentage of B220+, CD3+ and myeloid cells among donor-derived cells in peripheral blood 24 weeks after transplant. *P < 0.05; shown are mean values + 1 s.e. Mice were considered as engrafted when the percentage of Ly5.2+ cells in peripheral blood was higher than 1.0 and contribution was detected for all peripheral blood lineages. Data are based on two different lentiviral infection/transplant experiments with 5–7 recipient mice per group (for example, n = 10). h, Ratio of the densitometric score of the total Cdc42 expression as shown in Fig. 4d. The experiment was repeated four times with mice (1 mouse for 1 sample) from different lentiviral infection/transplant experiments. Shown are mean + 1 s.e., n = 4, *P < 0.05. i, j, Percentage of donor-derived LT-HSCs polarized for Cdc42 (i) and tubulin (j) 24 weeks after transplant. Shown are mean values + 1 s.e., n = 4, ∼200 cells scored per sample in total. *P < 0.05. k, Representative immunofluorescence z-stack pictures of tubulin (green) and β-catenin (red) localization in aged Wnt5a-NT (Ly5.2+GFP+) or aged Wnt5aKD (Ly5.2+GFP+) LT-HSCs. Nuclei are stained with DAPI (blue). Shown is also the final three-dimensional reconstructed merged image. Scale bar, 5 μm.
Extended Data Figure 5 Wnt pathways in HSCs and ageing.
a–g, Reverse transcriptase real-time PCR analysis of Notch1, Notch2, Jag1, Jag2, Dll1 (delta like1), Dll4 (delta like 4) and Hes1 transcript levels in young, aged and young Wnt5a-treated (16 h treatment) LT-HSCs. Notch3, Notch4 and Dll3 (delta like 3) transcripts were below detection limits in all the assayed samples. Data are expressed as fold difference compared to the expression in young LT-HSCs arbitrarily set to 1. Data were analysed with the 2−ΔΔCt method and plotted on a logarithmic or linear scale. Bars are mean + 1 s.e.; n = 3, *P < 0.05. h, Representative immunofluorescence picture of p-CamKII (green) expression and localization in young control and young Wnt5a-treated LT-HSCs. Pictures are shown on a dark background and as overlap with DAPI (staining nuclei). Scale bar, 5 μm. i, Relative expression of p-CamKII in young LT-HSCs and on Wnt5a treatment, determined by integration of pixel intensity. *P < 0.05. j, Representative immunofluorescence picture of NFATc (green) expression and localization in young control and young Wnt5a-treated LT-HSCs. Pictures are shown on a dark background and as overlap with DAPI (staining nuclei). Scale bar, 5 μm. k, Relative expression of NAFTc in young LT-HSCs and on Wnt5a treatment, determined by integration of pixel intensity. *P < 0.05. l, Changes in intracellular Ca2+ concentrations in ST-HSCs and LT-HSCs in response to stimulation with Wnt5a as determined by flow cytometry. m, Reverse transcriptase real-time PCR analysis of p57 and p27 transcript levels in young, aged and young Wnt5a-treated (100 ng ml−1, 16 h treatment) LT-HSCs. Data are expressed as fold difference compared to the expression in young LT-HSCs arbitrarily set to 1. Data were analysed with the 2−ΔΔCt method and plotted on a logarithmic or linear scale. Bars are mean + 1 s.e.; n = 3, *P < 0.05.
Extended Data Figure 6 Mechanisms of haematopoietic stem-cell ageing.
Cartoon scheme summarizing the main phenotypic and functional differences between young and aged LT-HSCs.
Supplementary information
Wnt5a is highly expressed in aged LT-HSCs
Confocal 3-D reconstruction of an aged LT-HSC stained for Wnt5a (green), Clathrin (red) and the nucleus (DAPI, blue). The cell was scanned along the z-axis to acquire a picture of the xy-plane every 0.6µm. (MP4 638 kb)
β-catenin is highly expressed and localizes mainly to the nucleus in young LT-HSCs
Confocal 3-D reconstruction of a young LT-HSC stained for β-catenin (red) and tubulin (green). The nucleus is stained with DAPI (blue). The cell was scanned along the z-axis to acquire a picture of the xy-plane every 0.6µm. (MP4 1288 kb)
β-catenin is very low expressed and localizes mainly outside the nucleus in aged LT-HSCs
Confocal 3-d reconstruction of an aged LT-HSC stained for β-catenin (red) and tubulin (green). The nucleus is stained with DAPI (blue). The cell was scanned along the z-axis to acquire a picture of the xy-plane every 0.6µm. (MP4 496 kb)
Rights and permissions
About this article
Cite this article
Florian, M., Nattamai, K., Dörr, K. et al. A canonical to non-canonical Wnt signalling switch in haematopoietic stem-cell ageing. Nature 503, 392–396 (2013). https://doi.org/10.1038/nature12631
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature12631
This article is cited by
-
WNT Signaling in Stem Cells: A Look into the Non-Canonical Pathway
Stem Cell Reviews and Reports (2024)
-
Childhood hematopoietic stem cells constitute the permissive window for RUNX1-ETO leukemogenesis
International Journal of Hematology (2023)
-
The Mechanism of Stem Cell Aging
Stem Cell Reviews and Reports (2022)
-
Noncanonical Wnt5a signaling regulates tendon stem/progenitor cells senescence
Stem Cell Research & Therapy (2021)
-
Kidney and epigenetic mechanisms of salt-sensitive hypertension
Nature Reviews Nephrology (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.