Abstract
The community of cells lining our airways plays a collaborative role in the preservation of immune homeostasis in the lung and provides protection from the pathogens and pollutants in the air we breathe. In addition to its structural attributes that provide effective mucociliary clearance of the lower airspace, the airway epithelium is an immunologically active barrier surface that senses changes in the airway environment and interacts with resident and recruited immune cells. Single-cell RNA-sequencing is illuminating the cellular heterogeneity that exists in the airway wall and has identified novel cell populations with unique molecular signatures, trajectories of differentiation and diverse functions in health and disease. In this Review, we discuss how our view of the airway epithelial landscape has evolved with the advent of transcriptomic approaches to cellular phenotyping, with a focus on epithelial interactions with the local neuronal and immune systems.
Similar content being viewed by others
Introduction
It is now well accepted that the cells lining the airways constitute more than just a barrier between the external environment and the underlying mesenchyme. This collection of specialized epithelial cells responds to microbes and noxious stimuli that overcome the mucociliary barrier and are a vital component of host defence, interacting with cells of the immune system to maintain homeostasis while facilitating immune reactions when necessary1. The respiratory epithelium must also manage responses to the diverse toxins contained within the inhaled environment and there is emerging evidence that epithelial dysfunction is a driver of numerous chronic diseases affecting the lungs. The traditional view of the epithelial layer incorporates basal cells in close proximity to secretory and ciliated cells, forming a tight unit that maintains a physical barrier but is also responsive to the inhaled environment via cells and molecules from the immune system. However, with the advent of advanced sequencing techniques, this view has changed to one of a dynamic cellular structure encompassing a wide range of highly specialized cells that are able to respond to environmental change, interact with resident microbial communities and cooperate with multiple other specialized cellular systems such as the immune and neural systems.
The respiratory tract is a complex organ system divided into the upper respiratory tract, that includes the nasal cavity, pharynx and larynx, and the lower respiratory tract comprising the conducting airways (trachea, bronchi and bronchioles) and the respiratory zone (respiratory bronchioles and alveoli) (Fig. 1). Each area has a specific function and the regional differences in cellular composition reflect this. Specialized epithelial cell populations line the entire respiratory tract from the nasal cavity to the alveoli. Elegant electron microscopy studies provided early insight into the morphology and ultrastructure of the principle epithelial cell types residing in the human airways2,3,4. In addition to using classical electron microscopy-defined morphological features, standard immunohistochemical staining for cell type-specific markers has been used to characterize and quantify epithelial cell populations throughout the human respiratory tract, determining the influence of anatomical location on cellular composition5,6,7. The airways are lined by ciliated and secretory cells primarily adapted to facilitate mucociliary clearance of particulate matter and infectious pathogens in the air we breathe. Like other mucosal surfaces, the airway epithelium is at an interface with the environment and is therefore critically important to host defence. Despite sharing the same embryological origin as the gut mucosa, immunological activity at the airway mucosal surface is necessarily distinct and shaped by differences in environmental conditions (temperature gradient, bidirectional airflow), resident microbial communities and airborne antigens8. Although not the focus of this Review, the epithelial cells lining the distal alveolar region of the lung are phenotypically and functionally distinct; alveolar epithelial type 1 (AT1) cells provide a specialized surface for gas exchange and type 2 (AT2) cells secrete pulmonary surfactant to prevent alveolar collapse during expiration. In each anatomical niche, there are progenitor cell populations, for example, basal cells in the airways and AT2 cells in the alveoli, that ensure robust epithelial regeneration under homeostatic conditions and following injury9.
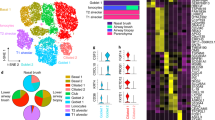
The traditional view of the airway epithelium — comprising epithelial cells lining the upper and lower respiratory tract — has been transformed by single-cell RNA sequencing (scRNA-seq). The revised contemporary landscape features newly identified cell types, such as the ionocyte, and different cell states. The cellular composition and cell states within the airway varies according to anatomical location (proximal–distal axis) and the presence of disease. The approach used to obtain human samples for sequencing differs according to lung compartment; the principle methods for sampling human airway epithelial cells are bronchial brushings and endobronchial biopsies conducted during bronchoscopy, in contrast to sampling of the alveolar region, which is achieved using parenchymal lung tissue obtained from surgical biopsy or from explants. BAL, bronchoalveolar lavage.
The advent of novel sequencing techniques has facilitated not only the identification of novel cell types but reveals potential functions of previously named but poorly understood cell types, for example, tuft/brush cells. There is also an indication that heterogenous cell types and states exist and that subtle changes in these will be influenced by the changing environment, for example, by smoking or exposure to allergens or pollutants. It is now clear that the airway wall represents a dynamic ‘community’ of epithelial cells existing in close association with resident immune and neuronal cells to generate an integrated unit that plays a critical role in maintaining mucosal immune homeostasis as well as facilitating host defence against inhaled pathogens. In this Review, we outline how single-cell transcriptomic techniques have been used to map the airway epithelial landscape and how this has highlighted the function of novel specialized epithelial cells and how they interact with cells of the immune and neuronal systems to regulate airway immunity.
Mapping the airway epithelial landscape
Single-cell RNA-sequencing (scRNA-seq) has transformed our view of the airway epithelium, unveiling a level of cellular diversity that had not been documented using microscopy for phenotyping10. This technology profiles the transcriptome of individual cells and therefore facilitates an unbiased characterization of different cell subsets in a heterogenous cell population, driving the discovery of unidentified cell types and states11,12,13. The Human Cell Atlas Consortium aims to comprehensively chart — at single-cell resolution with integration of spatial data — the changes that occur in lung cell composition and molecular phenotype in health and disease14. Added to this, it is also possible to computationally interrogate epithelial–immune interactions in the airway niche using known ligand–receptor interactions10,15,16,17.
Studying human epithelial biology using single-cell transcriptomics relies on the availability of lung tissue. One of the earliest studies in humans analysed diseased explant parenchymal lung tissue from patients with idiopathic pulmonary fibrosis (IPF) and lung lobes from donors unsuitable for transplant as control tissue18. For studies of the human airway epithelium, the acquisition of bronchial brushings and endobronchial biopsy samples in the setting of disease can be achieved through fibreoptic bronchoscopies carried out in the context of routine clinical care10,19. It is important to note that the cellular content differs by the sampling method used and this will influence the cell clusters obtained by scRNA-seq (Fig. 2). Acquiring age-matched, healthy control samples is more challenging as it necessitates research volunteers undergoing an invasive procedure. The procurement of appropriate human lung tissue is only feasible through close collaboration between clinicians and scientists and through human tissue biobanks. Importantly, logistical practicalities require careful consideration because the hypothermic preservation of tissue and the minimization of time delays between fresh sample collection in the clinic and processing in the laboratory are critical to ensuring high-quality scRNA-seq data. A cold ischaemic time of 72 h results in an increase in the presence of mitochondrial reads due to cellular stress or death in multiple lung cell types and a cell-specific decrease in the proportion of CD4+ and CD8+ cytotoxic T cells20. The enzymatic dissociation of tissue with collagenase is widely used to isolate cells for scRNA-seq studies but, as a caveat, it can induce the expression of immediate early genes, including Fos and Jun, and heat shock protein (HSP) genes in a subpopulation of cells, thus also potentially introducing an important artefact and potential bias in single-cell data sets21.
For human single-cell RNA sequencing (scRNA-seq) studies of the lower respiratory tract, a flexible bronchoscopy is performed to obtain bronchoalveolar lavage (BAL) fluid, airway wall brushings and endobronchial biopsies. The starting material determines the relative contributions of airway immune cells, epithelial cells and underlying stromal cells that are sequenced. A viable, single-cell suspension is obtained by enzymatic dissociation of brushings and biopsies. Bioinformatic pipelines generate distinct cell clusters by dimensional reduction and visualization techniques such as uniform manifold approximation and projection (UMAP) and facilitate assignment of cluster annotations based on cell-marker genes. Novel cell clusters and cell states can be determined in health and disease and trajectory analysis can be used to study dynamic, differentiating cell types.
During scRNA-seq analysis, cells sharing similar gene expression profiles are clustered together and can be assigned an annotation based on known marker genes11,13 (Table 1). In the first single-cell studies of the mammalian airway, a new cluster of cells was identified that did not map to any of the previously known epithelial cell types and, based on its gene expression signature, was termed the ‘pulmonary ionocyte’22,23. This sentinel discovery of a novel cell type highlights the power of scRNA-seq (Boxes 1,2).
Sub-clustering can be used to identify diverse cell states within a specific cell-type cluster. A single-cell analysis of human nasal epithelial cells differentiated in air–liquid interface cultures demonstrated a precursor subset of FOXJ1-expressing multiciliated cells termed ‘deutrosomal cells’, which express specific genes, including DEUP1 (also known as CCDC67), FOXN4 and CDC20B, all of which are critical for centriole amplification and ciliogenesis24,25. Deutrosomal cells are identifiable in the healthy human airways by scRNA-seq19.
Subtle but distinct molecular states of basal, goblet and ciliated cells denoted by differential marker gene expression have now been described in the human airway10. Goblet cells could be divided into two subsets according to their uniquely expressed genes; for example, ‘goblet 2’ cells in the nasal epithelium have higher expression of genes involved in immune cell recruitment such as CXCL10, IL19 and CSF3 (ref.10). The anatomical origin of the cell population within the respiratory tract plays an important role in governing its transcriptional signature and cell state. A single-cell study of brushings and biopsies taken from four distinct anatomical locations in healthy subjects, highlighted region-specific sub-clusters of suprabasal, secretory and ciliated cells between nasal and tracheobronchial compartments19. Spatial variations in gene expression profiles between cells of the same type relate, in part, to the biological function required at a specific airway location. Secretory cells in the nasal epithelium are enriched in gene sets associated with cell motility, differentiation and sensory perception pathways, whereas in the bronchial tree, they are enriched in genes related to innate immunity and wound healing response pathways19. Noteworthy is the inconsistent annotation of secretory cell types, including club cells and goblet cells, between single-cell studies due to overlapping gene expression signatures10,19,26.
A further determinant of airway epithelial cell state is revealed by the comparison of cells from healthy volunteers versus those collected from patients with respiratory disease. An analysis of airway wall biopsies from mild-to-moderate asthmatics exposed several cell states not found in the healthy airway10. Cells co-expressing gene markers of ciliated cells, including FOXJ1, and goblet cell genes such as MUC5AC, were termed ‘mucous ciliated cells’ and are postulated to represent a novel transitionary cell state driven by IL-4/IL-13 signalling in asthma.
Single-cell analyses of parenchymal lung tissue from patients with IPF, a progressive scarring lung disease, have reported a previously unidentified, transcriptionally distinct cluster of cells in the fibrotic distal lung that express some but not all markers of airway basal cells27,28. These intriguing ‘aberrant basaloid’ cells, which are KRT5–KRT17+, co-express genes encoding mesenchymal markers such as collagen type 1 α1 chain (COL1A1), the transcription factor SOX9 (which plays a role in distal airway development) and genes linked to IPF pathogenesis, including MMP7, which encodes matrilysin.
The pulmonary epithelium represents a dynamic cellular community and single-cell transcriptomics captures a unique snapshot of cells at different stages in the continuous process of differentiation — a feature that was first harnessed to elucidate epithelial lineage hierarchies at four distinct stages of alveolar development in mice29. With the refinement of scRNA-seq bioinformatic approaches it has been possible to study cell fate in human samples and to unveil the subtleties of human airway epithelial cell differentiation10,24,30. The transcriptional changes that accompany the differentiation of each cell can be exploited computationally by trajectory inference algorithms that order cells according to progress along their individual trajectories, which is quantified by ‘pseudotime’31,32. A pseudotime analysis of nasal airway epithelial cells in air–liquid interface cultures demonstrated that goblet cells could act as precursors to ciliated cells and that transitory FOXJ1+MUC5AC+ cells could be found in the context of this differentiation trajectory24. Furthermore, leveraging the relative abundance of unspliced and spliced mRNA per gene, it is possible to estimate the rate of gene expression change, termed ‘RNA velocity’, and to predict future cell states33,34.
Pseudotemporal ordering of cells using a diffusion map approach35 identified an intriguing population of Krt13-expressing cells in the mouse airway as an intermediary in basal to club cell differentiation23. This transitional cell was arranged in discrete, stratified, high-turnover structures termed ‘hillocks’ and expressed genes associated with squamous epithelial differentiation, cell adhesion and immunomodulation.
The comprehensive transcriptomic information charted at a cellular level by scRNA-seq has heightened our knowledge of the airway epithelial landscape at steady state. This technology has also been harnessed to study airway samples from critically ill patients with COVID-19 in order to provide finer grain insight into the pathogenesis of severe acute respiratory system coronavirus 2 (SARS-CoV-2) infection16,36,37. Studying single-cell gene expression in multiple tissue types from data sets collated from the Human Cell Atlas Consortium, genes associated with SARS-CoV-2 cell entry, namely ACE2, which encodes a viral entry receptor, and TMPRSS2, were significantly enriched in nasal secretory and ciliated epithelial cells. Noteworthy was the high ACE2 expression in cells at other mucosal surfaces, including the cornea, conjunctiva, small intestine and colon, which may, in combination with high nasal expression, explain the higher transmissibility of SARS-CoV-2 (ref.36). This demonstrates how existing data sets can be exploited to further our understanding of the pathogenesis of a novel emerging disease.
Functions of rare epithelial cell types
Basal cells are multipotent stem cells that account for a third of all airway epithelial cells and give rise to other major subpopulations, such as secretory and ciliated cells, during homeostatic maintenance of the epithelial barrier and regeneration in response to injury38. Basal cells are anchored to the basement membrane of the conducting airways and express keratin 5 (KRT5) and transcription factor tumour protein p63 (TP63). Lineage tracing and single-cell transcriptomic studies have demonstrated that functionally distinct basal cell subsets exist in the airway24,39. Two discrete basal cell states are present in the upper and lower airways, but their frequency is lower in the upper airways. These cell states reflect distinct differentiation stages in basal cells, with mature basal cells (located at a more apical region) expressing lower levels of TP63 and NPPC than the less mature basal cells10. Basal cells with a lower TP63 expression and higher KRT19 and NOTCH3 expression were annotated as ‘suprabasal’ cells by Deprez et al.19.
In addition to these major cell types, rarer cell populations comprising less than 1% of all human airway epithelial cells have been identified via detailed single-cell transcriptomic analyses19,26. Although uncommon, these cells seem to have very distinct phenotypes and perform specific tasks that are vital for the efficient functioning and maintenance of the epithelial landscape.
Hillock cells
The advent of scRNA-seq has enabled investigators to examine cell development trajectories. In addition to the connection between basal, club and ciliated cells, Montoro et al. defined a distinct trajectory, which links tracheal basal and club cells via a novel transitional cell that uniquely expresses Krt13 and Krt4 (ref.23). By looking at the location of these cells within the lungs coupled with lineage tracing in mice, the authors noted that Krt13+ cells lie within contiguous groups of stratified cells that do not contain luminal ciliated cells. They termed these groupings ‘hillocks’, a structure composed of luminal Scgb1a1+Krt13+ cells lying on top of Trp63+Krt13+ basal cells, with the expression of Trp63 occurring along a gradient from basal to suprabasal divisions. Critically, hillocks contain a particularly high number of cycling cells and expressed markers of cellular adhesion and epithelial differentiation as well as genes associated with barrier function and immunomodulation23. A parallel study by Plasschaert et al. confirmed the presence of a distinct population of Krt13+Krt4+ cells in the murine trachea and in ex vivo differentiated human basal epithelial cells, although they concluded that this population represented an intermediate population between basal stem cells and differentiated luminal secretory cells22. Furthermore, Krt13+Krt4+-expressing cells emerged in the tracheal epithelium 1 day after polidocanol-induced injury in mice22. In humans, KRT13 is expressed in the airway epithelium by a subpopulation of ‘cycling’ basal cells that are characterized by the expression of genes associated with cell proliferation, including MKI67 (which encodes Ki67)10. Further analyses are needed to confirm whether hillocks represent a truly distinct niche rather than a metaplastic zone in transition and to determine their origin and purpose. Indeed, it is not clear whether these structures are present throughout the airways or are restricted to the trachea. In support of their existence, Deprez et al. describe a population of KRT13+ cells, a high proportion of which were cycling, in nasal turbinates, indicating that hillock cells may exist in other areas of the human respiratory tract19.
Tuft cells
The respiratory tract contains several groups of chemosensory epithelial cells that coordinate interactions with the external environment — the enteroendocrine cells and the tuft cells (also known as ‘brush’ cells). These chemosensory epithelial cells share similarities with taste cells and are predicted to evoke both positive and negative responses from immune and neuronal cells40. They have a distinctive morphology, being bottle shaped with apical microvilli, and are expressed in a range of organs, including the gut and airways. Their existence is well documented in the gut, where they are thought to be triggered by dietary metabolites such as succinate and bacterial products (for example, quorum-sensing lactones or d-amino acids)41,42,43. By contrast, their role in the lung is less certain, but they have been detected in the nose, trachea and proximal airways and exist in close contact with nerve fibres, mediating communication between neuronal and immune pathways. However, they are not universally identifiable; indeed, Vieira Braga et al.10 could not identify a unique population of tuft cells in a scRNA-seq analysis of the human airways using both bronchial brushing and endobronchial biopsy. This likely reflects the rarity of these cells and perhaps the methods used for the collection of airway material.
Tuft cells express a range of indicative markers, including POU domain, class 2, transcription factor 3 (POU2F3), which is considered the lineage-defining transcription factor for tuft cell specification40, as well as transient receptor potential cation channel subfamily M member 5 (TRPM5). scRNA-seq has now identified two terminally differentiated Trpm5+ tuft cell populations; one is positive for Gng13 and is likely to be responsible for ‘taste’ sensing, and the other is positive for Alox5ap, suggesting that it contributes to leukotriene synthesis22,23. Tuft cells are thought to generate cysteinyl leukotrienes via the ATP sensor P2Y2 (ref.44). Although generally present in low numbers, the expansion of tuft cells in the airways occurs following the inhalation of common aeroallergens via activation of the cysteinyl leukotriene pathway. Endogenously generated leukotriene E4 is sensed via CysLT3R, which regulates the number and function of murine airway brush cells45.
Thus, tuft cells are able to respond to a diverse range of signals via the combinatorial expression of different receptors. Tuft cells are thought to promote protective respiratory reflexes, such as sneezing, but can also contribute to apnoea as well as to local neurogenic inflammation of the mucosa46,47. Given their distribution and function, tuft cells have also been referred to as ‘solitary chemosensory cells’, enhancing the chemoresponsive protection provided by the local neuronal system. scRNA-seq analysis shows that mouse tuft cells express high levels of cholinergic and bitter taste signalling transcripts (Tas2r108, Gnat3, Trpm5). Acetylcholine is released from tracheal tuft cells following stimulation with either the bitter taste receptor type 2 (TAS2R) ligand denatonium or Pseudomonas quinolone quorum-sensing molecules, increasing mucociliary clearance mediated by TRPM5 and M3 muscarinic acetylcholine receptor (M3R), together with calcium release from ciliated cells48. It is postulated that the detection of these quorum-sensing molecules from Gram-negative pathogenic bacteria offers a mechanism by which the epithelium triggers capsaicin-sensitive nerve fibres, releasing calcitonin gene-related peptide (CGRP) and substance P, which promote innate immune cascades and microvascular leak and combat bacterial invasion, thus limiting the population densities capable of forming destructive biofilms43.
Solitary chemosensory cells, with tuft cell properties, are found to be the primary epithelial source of IL-25 in patients with chronic rhinosinusitis with nasal polyps, facilitating the activation of group 2 innate lymphoid cells (ILC2s) and the production of IL-13, thereby maintaining the type 2 environment in the upper airways49. Once activated, tuft cells can release neurotransmitters (such as acetylcholine), eicosanoids involved in leukotriene and prostaglandin biosynthesis and ATP, and cytokines such as IL-25 and thymic stromal lymphopoietin (TSLP)40,42. All of these mediators are implicated in allergic inflammation, being able to elicit ILC2 activation and accumulation via the expression of specific receptors50.
Ionocytes
Single-cell transcriptomic studies of the mouse tracheal epithelium and differentiated human tracheal epithelial cells in culture discovered a novel cluster of cells, termed ‘pulmonary ionocytes’ due to the similarity of their gene expression profiles to those of the ion transport cells of the mucociliary epithelium of Xenopus larval skin22,23,51. As in Xenopus, ionocytes express genes encoding subunits of a V-ATPase proton pump, which regulates ion transport and pH. Crucial in determining the ionocyte lineage in mouse and human epithelial cells is FOXI1, which belongs to the forkhead family of transcription factors. Knockout studies in mice and lentiviral studies in human cell cultures show that FOXI1 regulates the expression of the CFTR gene that encodes a critical chloride-ion transporter that is defective or absent in cystic fibrosis. Cystic fibrosis is a life-limiting disease characterized by increased mucus viscosity, impaired mucociliary clearance, chronic infection and airway inflammation, leading to loss of lung function52. Ionocytes account for only 1–2% of human airway epithelial cells but are more enriched in CFTR mRNA than in any other airway cell type. Combining scRNA-seq with conventional in vivo lineage tracing over three different time points in a technique called ‘pulse-seq’, indicated that basal cells were the principal source of ionocytes and other rare cell types23. Given the clustering of rare cell types in scRNA-seq data sets, Goldfarbmuren et al. further investigated rare cell lineage relationships using a CRISPR–Cas9 knockout system in human tracheal basal cells to determine that, during differentiation, ionocytes and pulmonary neuroendocrine cells (PNECs) arise from POU2F3+ tuft-like cells26.
Pulmonary neuroendocrine cells
PNECs are solitary cells resident within the surface epithelium of the trachea, bronchi and bronchioles. They can also exist in formations called neuroendocrine bodies in the intrapulmonary airways. PNECs function as chemosensors of the airway and respond to changes in oxygen, stretch and chemical stimuli53. They are a rich source of neuropeptides and neurotransmitters that elicit immune and physiological effects. PNECs are the only innervated airway epithelial cell type, are conserved across species and represent just 1% of the total lung epithelial cell population. They are reported to be the earliest specialized cell type that forms in the lung epithelium during development53,54,55 and are thought to be particularly important functionally in early life since neonatal mice lacking PNECs are protected from allergic inflammation56. In vitro analyses predicted roles for PNECs in a range of activities, including oxygen sensing, the maintenance of bronchial and vascular smooth muscle tone, and in the coordination of immune responses. However, an elegant series of experiments with mutant mice has confirmed the functional characteristics of PNECs that influence a range of pulmonary diseases. A defining characteristic marker of PNECs is the gene Ascl1, a transcription factor that is essential for their formation. Mice that lack Ascl1 die at birth, but a conditional knockout designed to inactivate Ascl1 in PNEC precursors led to viable mice that completely lack PNECs, as marked by a lack of CGRP+ cells in the airway epithelium56. These Ascl1-mutant mice were normal at baseline but were protected from developing severe goblet cell hyperplasia and type 2 inflammation when exposed to allergens during the perinatal period. This absence of PNECs was accompanied by a reduction in key neuropeptides, including the neurotransmitter GABA.
The expression of the roundabout (ROBO) genes is vital for the clustering of PNECs into neuroendocrine bodies, limiting immune cell infiltration, and in preventing alveolar simplification during postnatal lung development. Although PNEC defects were apparent in Robo-deficient mice at E15.5, the physiological effects only occurred after birth, indicating that the effects of PNEC are dependent on the physical exposure to air that occurs with the first breath, thus reinforcing the idea that PNECs are sensors of the changing inhaled environment. By contrast, the genetic ablation of PNECs in adult mice had no effect on airway homeostasis or repair following chemical injury.
In early life murine models, PNECs were shown to facilitate mucus hypersecretion during allergen exposure via neurotrophin 4 regulation of PNEC innervation and the secretion of GABA, which promotes Muc5ac expression. PNECs represent the only source of GABA in primate lungs and in ex vivo cultures of human epithelial cells57,58. PNECs also reside in close association with ILC2s in the airways and communicate via CGRP to maximize the ILC expression of IL-5 and GABA to elicit mucus production56,59. GABA production was found to be absolutely required for goblet cell hyperplasia but not in type 2 inflammation in both neonatal and adult models of allergic inflammation56. Importantly, these findings are reflected in humans, with increased numbers of the CGRP+ subset of PNECs in patients with allergic asthma.
The fact that abnormalities in PNEC numbers are associated with a wide range of pulmonary diseases, including rare genetic disorders such as congenital diaphragmatic hernia and small cell lung cancer as well as more common diseases such as asthma, indicate a key role for these cells in physiological and immune pathways critical for effective functioning of the lung at homeostasis.
Immune properties of airway epithelial cells
Antimicrobial mediators and airway mucins, including MUC5AC and MUC5B, are produced by secretory epithelial cells lining the airways and submucosal glands and contribute to the first layer of host defence at the airway epithelial surface60. Secretory IgA (SIgA) — produced by sub-epithelial plasma cells and transported to the apical surface of airway epithelial cells via the polymeric immunoglobulin receptor (pIgR) — prevents the adherence of airborne microorganisms in a process called ‘immune exclusion’61,62. Epithelial cells are equipped with pattern recognition receptors, such as Toll-like receptors, which rapidly sense and initiate an immune response to microbial threats, and cytokine receptors, including TNFR1, which allow them to respond to signals produced by immune cells such as airway macrophages63. Adjacent airway epithelial cells are linked by intracellular tight junctions and adherens junctions, which selectively regulate the paracellular diffusion of ions and molecules and maintain barrier integrity64. These tight junctions separate ligands present at the apical epithelial surface, for example, growth factor heregulin, from its ErbB family receptors located at the basolateral surface, so that activation occurs following injury and disruption of epithelial integrity65. Our view of the function of this mucosal surface has profoundly shifted because of studies demonstrating that airway epithelial cells possess intricate and sophisticated properties that allow them to direct host immunity, inflammation and remodelling66,67 (Fig. 3).
In addition to orchestrating local immune cells through the release of soluble mediators and cell–cell contact, epithelial cells possess intrinsic immune capabilities that directly influence barrier immunity. These include sensing of apoptotic cells by AXL receptor66 and clearance of apoptotic cells by RAC1 (ref.70) (part a), inflammatory memory67 (part b) and circadian control of inflammation76,79 (part c). BMAL1, brain and muscle ARNT-like 1; CXCL5, CXC-chemokine ligand 5; GAS6, growth arrest-specific protein 6; TH2, T helper 2; TGFβ, transforming growth factor-β.
Sensing and clearing apoptotic cells
The airway is an environment exposed to pollutants, pathogens and allergens known to induce apoptosis68. The clearance of apoptotic cells is carried out by ‘professional’ phagocytes, such as macrophages and dendritic cells69, but also by ‘non-professional’ phagocytes, including airway epithelial cells70. Apoptotic epithelial cells labelled with a CypHer5 dye that fluoresces within acidic phagolysosomes were directly engulfed through the recognition of phosphatidylserine by human bronchial epithelial cells (from a BEAS-2B cell line)70. The inducible deletion of the small GTPase RAC1, which is a downstream signalling molecule in the phagocytic engulfment pathway, in the airway epithelium using a Ccsp-Cre/Rac1fl/fl mouse model resulted in defective apoptotic cell phagocytosis and in a significant reduction in the production of the anti-inflammatory cytokines transforming growth factor-β (TGFβ) and IL-10 (ref.70). Critically, this translated into an exaggerated allergic airway inflammatory response in RAC1-deficient mice, with elevated levels of epithelial cell-derived IL-33 detected in bronchoalveolar lavage (BAL) fluid70. This data supports the concept that airway epithelial cell-mediated phagocytosis of apoptotic cells via RAC1 signalling plays a key role in taming the inflammatory response to common inhaled allergens. This study used a Ccsp-Cre mouse strain that specifically targets club cells and therefore this mechanism may not be generalizable to other types of airway epithelial cells.
The recognition of apoptotic cells by airway basal cells also plays a key role in determining cell phenotype and fate in the context of inflammation66. Basal cell hyperplasia is an early event in the pathogenesis of chronic obstructive pulmonary disease (COPD), an inflammatory lung disease71. The mechanism driving basal cell proliferation in the context of airway inflammation highlights a novel and previously unrecognized capability of basal cells. Under homeostatic conditions, mouse tracheal basal cells express the AXL receptor (from the TAM receptor tyrosine kinase family) bound to GAS6 (a bridging molecule)66. In a mouse model of H1N1/PR8 influenza A virus infection, which is characterized by airway inflammation and ciliated epithelial cell apoptosis, basal cell re-entry into the cell cycle and their proliferation was promoted by the presence of AXL. Similarly, apoptotic thymocytes delivered intranasally to C57BL/6 mice increased the number of Ki67-expressing proliferating basal cells, an effect that was significantly reduced in Axl-knockout mice. Therefore, it has been inferred that basal cell proliferation during airway inflammation is mediated through the recognition of apoptotic cells by the AXL receptor tyrosine kinase66. In the absence of AXL, basal cells may differentiate into other cell types by so-called asymmetric cell division to accelerate re-epithelialization. Immunostaining of the small airway epithelium from human COPD tissue samples revealed that the number of AXL-expressing basal cells was increased and that their proliferation correlated with the presence of caspase 3-positive apoptotic cells66.
Inflammatory memory
Not only are basal cells able to sense and respond to changes in the inflammatory microenvironment, but they also have an intrinsic capacity for inflammatory memory72,73 — a discovery that was made in the context of a chronic allergic inflammatory disease67. Chronic rhinosinusitis(CRS) is a type 2 immune-mediated disease characterized by inflammation and epithelial dysfunction in the nose and paranasal sinuses, with or without the formation of abnormal tissue outgrowths called polyps74. Basal cell hyperplasia is a feature of tissue remodelling in this condition. Single-cell transcriptomics was used to study human tissue samples collected during ethmoid sinus surgery from subjects with CRS with and without polyps as well as nasal scrapings from the inferior turbinate of healthy subjects and from those with CRS and polyps67. The most striking disease-related changes to the transcriptomic state were seen in basal cell, differentiating/secretory cell and glandular cell populations. In the polyp samples, there was notable basal cell expansion at the expense of epithelial cell diversity. Basal cells upregulated IL-4-responsive and IL-13-responsive genes and displayed a failure to differentiate by pseudotime analysis. A combination of bulk RNA sequencing and transposase-accessible chromatin sequencing (ATAC-seq) showed the upregulation of transcription factors such as KLF5 and ATF3, known to maintain undifferentiated cell states, corresponding to enriched motifs in sorted polyp basal cells. These findings suggested that intrinsic changes at an epigenetic level may be responsible for the differences observed in polyp basal cell state and may be governed by the local type 2 inflammatory milieu. Bulk RNA sequencing of ex vivo-stimulated basal cells in submerged cultures revealed that IL-4 and IL-13 induced more than 10 times the number of genes in non-polyp basal cells than in polyp basal cells. Additionally, at baseline, the Wnt pathway activator CTNNB1 was expressed in polyp basal cells at levels only achievable in non-polyp basal cells through IL-4 and IL-13 stimulation, suggesting a ‘memory’ of in vivo exposure to type 2 cytokines by polyp basal cells. These data have profound implications for our understanding of allergic diseases given that efforts to treat these diseases are generally focused on manipulating cells of the immune system.
Circadian rhythm
For over a decade, it has been recognized that, in addition to neuro-hormonal control of the circadian rhythm by the suprachiasmatic nucleus in the brain, peripheral tissues and their cells, including airway epithelial cells, possess an intrinsic circadian clock responsible for rhythmic immune oscillations with the time of day75. In mouse and human lung tissue, the clock gene products CLOCK and PER2 were expressed in CCSP+ club cells75. Circadian oscillations, measured by the bioluminescence of PER2 in ex vivo mouse lung slices from transgenic mice expressing a Per2-luciferase fusion gene (Per2-Luc mice), could be ‘reset’ in response to glucocorticoids and were lost with the selective ablation of club cells. A notable time-of-day variation in inflammatory response, in particular the BAL neutrophil count, was observed in response to an aerosolized lipopolysaccharide (LPS) challenge in C57BL/6 mice76. The targeted deletion of the clock gene Bmal1 in club cells using Ccsp-Bmal1−/− mice augmented the duration of the neutrophilic inflammatory response to LPS challenge as well as to Streptococcus pneumoniae across the circadian cycle but without an increase in myeloperoxidase activity or a reduction in bacterial load76. Driving this pulmonary inflammatory response was the neutrophil chemoattractant CXCL5, which lost circadian regulation in the Bmal1 knockout.
REV-ERBα is a nuclear receptor that sits in a negative feedback loop downstream of core transcriptional activators of the molecular clock, namely CLOCK and BMAL1 (ref.77). REV-ERBα plays a key role in the circadian modulation of innate immune responses in myeloid cells and airway epithelial cells78,79. The targeted deletion of the DNA-binding domain of REV-ERBα in CCSP+ bronchial epithelial cells in mice led to the amplification of airway neutrophilic inflammation upon an aerosolized LPS challenge79. Again, this inflammatory response was mediated through the increased expression of CXCL5 at the transcript and protein level. Interestingly, the dual deletion of REV-ERBα and its paralog REV-ERBβ resulted in a further increase in neutrophilic inflammation upon LPS challenge in these mice as well as in unchallenged mice at steady state, suggesting that REV-ERB proteins play a homeostatic anti-inflammatory function79. Inflammatory responses to LPS in vivo were not impacted by the deletion of REV-ERBβ alone, emphasizing the leading role of REV-ERBα in this pathway.
A further understanding of the local circadian control of cell function may reveal why some patients experience ‘night-time’ asthma or cough.
Immune cell interactions at the epithelial surface
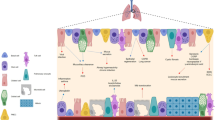
scRNA-seq facilitates the mapping of the diverse innate and adaptive immune cell communities of the airway niche at high resolution. Airway epithelial cells act in concert with resident and recruited immune cells to regulate pulmonary immunity (Fig. 4). Whilst transcriptomic studies of human sputum80 and BAL81,82 facilitate the detailed characterization of immune populations within the airway, bronchial brushing samples and biopsies capture both the epithelial and immune cell components10. Computational advances are now being leveraged to unveil the complex epithelial–immune crosstalk that occurs in the airway and alveolar space in health and disease10,83.
Complex crosstalk exists between epithelial cells and resident and recruited immune cell populations in the airways. These interactions depend on the specific environmental stimuli, for example, infection with viruses and bacteria or exposure to allergens. Transcriptomic studies have provided enhanced insight into the changes in cell functional state that are driven by these interactions. This figure provides specific examples and is not exhaustive. AHR, airway hyperresponsiveness; cDC2, type 2 conventional dendritic cell; CSF1, colony-stimulating factor 1; CXCL, CXC-chemokine ligand; CXCR6, CXC-chemokine receptor 6; DC, dendritic cell; IFN, interferon; LN, lymph node; MAIT cell, mucosal associated invariant T cell; TRM cell, tissue resident memory T cell.
Macrophages are the most abundant immune cells located in the airway lumen and are found readily in human sputum samples80,84, airway brushings10 and BAL82,85. Murine studies show that resident airway macrophages are perinatally derived from fetal monocytes and self-maintained locally throughout life86,87. This paradigm was challenged by recent human work revealing, through scRNA-seq, that following lung transplant, most tissue-resident airway macrophages in the donor lung were replaced by airway macrophages from the circulating monocyte pool of the recipient88. A single-cell transcriptomic study of sputum samples from patients with cystic fibrosis indicated a disease-related shift in dominant airway immune populations from resident airway macrophages to recruited monocytes and neutrophils, displaying pro-inflammatory phenotypes and gene expression changes suggestive of impaired phagocytosis80. Bi-directional interactions between resident airway macrophages and epithelial cells ensure the maintenance of a homeostatic state of immune tolerance to harmless stimuli and appropriate protective responses to inhaled pathogens with effective tissue repair when required89. A mouse model utilizing live confocal microscopy showed that CD11c-expressing alveolar macrophages formed direct connections with alveolar epithelial cells by connexin 43-containing gap junctions to limit the inflammatory response to LPS90. The exploration of cell–cell signalling networks using scRNA-seq data from lung tissue across four mammalian species (including humans) showed crosstalk between epithelial cells and macrophages in the alveolar cell niche at homeostasis83.
There are few studies that investigate epithelial–macrophage crosstalk in the human airways, yet this is a critically important area if we are to fully understand how dysregulated immunoregulatory interactions at this environmental interface contribute to human disease.
To explore the epithelial–immune axis in an inflammatory airways disease, BAL was studied in human subjects with atopic asthma before and after sub-segmental broncho-provocation with an identified allergen91. Colony-stimulating factor 1 (CSF1) was elevated in BAL samples after aeroallergen challenge and was shown to be secreted by airway epithelial cells. Epithelial cell-specific deletion of Csf1 in transgenic mice eliminated allergic airway inflammation. Furthermore, epithelial cell-derived CSF1 increased the numbers of a subset of alveolar dendritic cells expressing the CSFR1 receptor in BAL following allergen challenge and promoted the migration to regional lymph nodes91. Taken together, these findings suggest that epithelial cell–dendritic cell interactions in the airways enhance antigen presentation and augment adaptive allergic airway responses.
A robust immune response to respiratory pathogens at the airway epithelial surface is also contingent on an effective adaptive T cell response. Noteworthy is the induction of a subset of T cells — tissue-resident memory T (TRM) cells92 — at the site of antigen entry in a spatially restricted niche around the airways, poised to generate a rapid immune response to subsequently encountered pathogens93,94. CD4+ and CD8+ TRM cells expressing CD69 were transcriptionally distinct to circulating CD69– effector memory T cells and expressed adhesion molecules that promote retention within the lung mucosal tissue95. Moreover, parabiosis experiments in influenza-immune mice found that lung airway CD8+ TRM cells were continually replenished by interstitial CD8+ TRM cells rather than by circulating memory T cells96. This was mediated by chemokine receptor CXCR6 binding to CXCL16 secreted by airway epithelial cells and macrophages96. In human lung transplant recipients, donor TRM cells persisted in BAL samples from the lungs but not in peripheral blood for over 1 year97. A significant proportion of airway CD8+ TRM cells co-expressed the αE integrin CD103, which combines with β7 integrin to form the heterodimer αEβ7 and interacts with E-cadherin on epithelial cells98, promoting retention at this mucosal site. Donor and recipient T cells were localized around the airways in biopsy samples and infiltrating T cells from the recipient acquired a TRM phenotype over time in BAL samples97.
Crosstalk between TRM cells and airway epithelial cells can shape the immune response to environmental antigens. Pneumococcal pneumonia generates CD4+ TRM cells, which were discovered to fine-tune lung epithelial cells to increase CXCL5 expression and enhance neutrophil recruitment during heterotypic recall infections99. In the context of a house dust mite allergic airways mouse model, the persistence of CD4+ TRM cells around the airways was associated with a rapid response to allergen re-challenge, resulting in airway hyperresponsiveness, a cardinal feature of asthma100. Single-cell transcriptomics of airway wall biopsies from asthmatic patients and healthy controls defined two CD4+ T cell subsets — classic CD4+ TRM cells and a novel cluster called tissue migratory CD4+ T cells — that expressed genes associated with cell egression into tissues such as S1PR1 and SELL10. A bioinformatic tool was used in this study to interrogate cell–cell interactions in the asthmatic airway wall and unveiled increased predicted crosstalk between T helper 2 cells and epithelial cells, other immune cells, fibroblasts and smooth muscle cells known to play an important role in the pathogenesis of the disease.
The specific properties of the airway microenvironment can drive adaptive gene expression changes in immune cells by epigenetic programming, which shape cell phenotype and function101. Compared with TRM cells in the spleen and lung interstitium, following influenza infection, CD8+ airway TRM cells displayed a distinct transcriptomic and epigenetic profile, enriched for genes associated with the integrated stress response and with amino acid starvation, which ultimately led to a gradual loss of these cells over time by apoptosis101.
Spanning the innate and adaptive arms of the immune system, a subset of innate-like T lymphocytes called mucosal-associated invariant T (MAIT) cells, contribute approximately 4% of the total T cells in human airway wall biopsies102. The expression of a semi-invariant αβ T cell receptor (TCR) allows MAIT cells to recognize metabolites of riboflavin (vitamin B2) biosynthesis derived from bacteria and yeasts and presented by MHC-related protein 1 (MR1)103. When activated, MAIT cells generate a rapid pro-inflammatory cytokine response to provide protection against respiratory pathogens104 but also display a tissue repair transcriptomic signature, suggesting other important functions in barrier integrity and healing postinfection105.
Mediators produced by resident immune cells can hinder the epithelial cell response to pathogens at the airway mucosal surface. Type I (IFNα and IFNβ) and type III (IFNλ) interferons are key mediators in the host anti-viral response but have recently been shown to compromise epithelial repair following infection106,107. Sustained IFNλ produced by lung-resident dendritic cells in response to polyinosine:polycytidylic acid (poly(I:C)), a synthetic viral RNA analogue, impaired epithelial barrier integrity, leading to severe pathology with Staphylococcus aureus superinfection in a mouse model106. During the recovery phase of murine influenza infection, airway epithelial proliferation and regeneration was reduced in a TP53-dependent manner by prolonged exposure to IFNλ107. The nature of the epithelial interferon response is also directly influenced by the type of invading pathogen. A delayed type I interferon response drives the accumulation of inflammatory monocytes and macrophages and leads to severe immunopathology in mice infected with SARS-CoV108. Interestingly, the transcriptional response of human bronchial epithelial cells to SARS-CoV-2 was found to be distinct from that induced by other respiratory viruses — genes associated with type I and type III interferon pathways were suppressed, yet there was a robust expression of genes encoding chemokines and pro-inflammatory cytokines, which is likely to be responsible for many of the clinical manifestations of the disease109. Defects in the type I interferon response due to rare genetic variants or IgG neutralizing autoantibodies were also discovered in a proportion of patients with severe COVID-19 pneumonia110,111.
Complex interactions exist between epithelial cells and innate and adaptive immune cells in the airway to maintain host defence. Signals from epithelial cells maintain tissue resident immune cells and modulate their response to antigen while, conversely, immune cells can directly alter the function and phenotype of airway epithelial cells.
Airway neuroimmune interactions
The general location of airway epithelial cells enables them to sense and react to environmental change via the secretion of a range of mediators that facilitate interactions with immune and stromal cells in the underlying parenchyma. In addition to the specialized epithelial cells that monitor the airways and trigger avoidance reflexes, such as coughing and sneezing, it is well recognized that the airways are richly innervated with sensory neurons, facilitating the execution of these avoidance reflexes112. Recent evidence indicates that extensive crosstalk between neuronal and immune systems and specialized chemosensory pulmonary epithelial cells is vital for the regulation of tissue homeostasis113,114 (Fig. 5). This integrated landscape is also able to coordinate responses to invading pathogens or noxious particles, to monitor environmental changes such as fluctuations in temperature or oxygen, or even to react to physical events such as stretch or constriction that occur during gasping. For example, PIEZO1, a mechanosensory ion channel expressed by myeloid cells in mice, was shown to sense environmental cyclical hydrostatic pressure changes and generate a pro-inflammatory response115.
The lungs are densely innervated and complex interactions between epithelial cells, neurons and immune cells facilitate sensing of the external inhaled environment. Pulmonary reactions to stimuli, such as pathogens, pollutants, toxins and environmental changes, result in avoidance reflexes and inflammation. Sensory neurons release vasoactive peptide (VIP), which binds to VIP receptor 2 (VIPR2) expressed by group 2 innate lymphoid cells (ILCs) and T helper 2 cells, resulting in the release of IL-5, which stimulates the release of more IL-5 from these neurons, generating an amplifying loop to enhance allergic responses. Communication between group 2 ILCs and pulmonary neuroendocrine cells (PNECs) occurs via calcitonin gene-related peptide (CGRP) to maximize the ILC-derived expression of IL-5 and GABA to elicit mucus generation. By contrast, CGRP inhibits neutrophils and γδ T cells during bacterial infections. Cholinergic neurons react to allergens and helminths to amplify type 2 inflammation via the secretion of neuromedin U (NMU) which reacts with NMU receptor 1 (NMUR1) on group 2 ILCs to enhance cytokine secretion.
The lungs contain a range of different neural cells, including nociceptor sensory neurons that react to noxious or potentially harmful stimuli, while cholinergic neurons regulate airway tone, airway smooth muscle contraction, mucus secretion and vasodilation via the interaction with muscarinic acetylcholine receptors (mAChRs) found on airway smooth muscle, glands and pulmonary vasculature. Given that many basic respiratory responses, such as cough, are mediated by sensory neurons116, it is perhaps not surprising that they are predicted to play an important role in lung inflammatory responses. In addition to relaying sensory changes to the brain, activated lung neurons can themselves release stimulatory peptides that contribute to the ensuing inflammatory reaction. Direct evidence of their contribution to pulmonary inflammation stems from experiments in which the ablation or chemical suppression of Nav1.8+ or transient receptor potential vanilloid type 1 (TRPV1) sensory neurons resulted in reduced allergic inflammation and bronchial hyperresponsiveness117,118,119. Similarly, targeted lung denervation, a bronchoscopic radiofrequency ablation therapy, is used in patients to durably disrupt parasympathetic pulmonary nerves because it has been found to decrease airway resistance, mucus hypersecretion and inflammation117,120 in patients with COPD.
The combination of specialized chemosensory cells and direct neuronal interaction may also represent an alternative way in which the airways respond to inhaled respiratory pathogens, airborne pollutants and allergens. The activation of epithelial TRPV4 triggers protective responses to bacterial LPS, an increasing ciliary beat frequency and the production of bactericidal nitric oxide. Mice lacking TRPV4 display exacerbated airway hyperreactivity and enhanced neutrophil recruitment into the airways121. Conversely, the specific deletion of TRPV1+ neurons resulted in enhanced immune protection to lethal pneumonia induced by S. aureus via increased survival cytokine induction and bacterial clearance122. TRPV1 and Nav1.8+ nociceptor sensory neurons were shown to influence bacterial dissemination via the suppression of neutrophil numbers and surveillance activities and the regulation of resident γδT cell numbers. The specific ablation of vagal TRPV1 neurons resulted in enhanced antibacterial immunity via the release of the neuropeptide CGRP.
Epithelial–neuronal interactions facilitate lung inflammation via the secretion of neuropeptides and neurotransmitters. Many immune cells express receptors that facilitate communication with the epithelium but also directly with nerves123. The discovery that ILC2s express a range of receptors for neuropeptides and neurotransmitters reveals a mechanism by which neuronal and immune systems integrate to promote a range of type 2 cytokine responses that facilitate antimicrobial, inflammatory and tissue protective type 2 repair responses at mucosal sites124. ILC2 residency in the airways is regulated by signals delivered systemically or by the local tissue environment125 and these cells are enriched at branch points, similar to PNECs56. ILC2s interact directly with neurons via neuropeptides, neurotransmitters and neurotrophic factors, including CGRP, neuromedin U (NMU), acetylcholine and vasoactive intestinal peptide (VIP) and such interactions facilitate both stimulatory and inhibitory signals123. ILC2 expression of VIPR2 enables the cells to respond to neuronal-derived VIP by secreting IL-5, thus creating a feedback loop whereby IL-5 stimulates nociceptors to produce more VIP. NMUR1 signalling amplifies ILC2-driven allergic inflammation126. NMU secreted by thoracic dorsal root ganglia interacts with NMUR1 expressed by lung ILC2s and regulated by IL-25. The loss of NMUR1 reduces ILC2 responses and type 2 inflammation127. NMU also promotes the contraction of smooth muscle cells, although this has not been shown specifically in pulmonary smooth muscle cells128. Conversely, CGRP has differential effects on the immune response to allergen-induced inflammation. Mice lacking PNECs have a reduced expression of both CGRP and GABA after allergen exposure. CGRP induces IL-5 secretion from ILC2s and inactivation of the CGRP receptor (Calcr1) in ILC2s in vitro ameliorates allergic inflammation56. CGRP is also found to negatively regulate type 2 responses to either allergen exposure or helminth infection59,126. CGRP modulates type 2 cytokine production from ILC2s that have been activated by alarmins or NMU, thereby constraining the magnitude of the type 2 responses in vivo59. PNEC-derived GABA seems to affect mucus secretion since the inactivation of GABA production results in defective goblet cell hyperplasia and direct instillation of GABA in PNEC-deficient mice restores mucus production56. These differential effects of neuropeptides on ILC2 cytokine production and type 2 immune pathology, concomitant with epithelial cell-derived alarmins and neuropeptides, provides an integrated mechanism whereby tissue responses to allergen can be fine-tuned according to changing circumstances.
Concluding remarks and future directions
Our views of the human airway epithelium have shifted dramatically, catalysed by the advancement in technologies that facilitate single-cell transcriptomics. The cellular communities of the airway epithelium are diverse, dynamic and yet functionally cohesive. Epithelial cells work collaboratively with localized populations of immune cells to support a multifaceted frontline defence system with highly effective injury-repair responses.
Epithelial, stromal and immune cells in the lung can be modulated by many aspects of their local tissue microenvironment, including the properties of the extracellular matrix129,130, mechanical stress131 and exposures such as to cigarette smoke26,132. Mapping cell location in a specific anatomical tissue microenvironment using spatial transcriptomic techniques133 will undoubtedly enhance our understanding of cell molecular phenotype and function within a particular spatial niche.
A multi-omic approach utilizing single-cell transcriptomics and mass spectrometry-based proteomics was used to chart the molecular and cellular alterations that occur in the lung with ageing134. This work showed alterations in airway epithelial cell composition and lung extracellular matrix with increasing age. The application of a similar approach to human tissue samples in combination with comprehensive demographic and clinical information will allow us to link changes in cell biology with disease phenotype.
The functional validation of transcriptomic data that has been generated through complex bioinformatics requires robust in vitro culture systems135. The human ‘small-airway-on-a-chip’ was developed to recapitulate the airway tissue microenvironment and cell responses in health and disease, and represents a significant development in this area136. This system comprises a microfluidic device with a fully differentiated, mucociliary airway epithelium cultured adjacent to a microvascular endothelium in a channel exposed to media under flow. It has been used to model inflammatory airway diseases and study the interplay between epithelial cells and immune cells in vitro.
The airway epithelium is dynamic and continuously replenished by basal progenitor cells38,137. Single-cell transcriptomics has been used to elucidate airway basal cell differentiation and response to injury in vitro24. Fascinating mechanisms exist to preserve a healthy barrier in other stratified epithelial surfaces around the body such as the skin. A natural phenomenon of cell competition was observed to operate in a mouse model of skin development — in a mosaic epidermal tissue, so-called ‘winner’ progenitor cells induce apoptosis in neighbouring apoptotic ‘loser’ progenitor cells and eliminate them by engulfment138. Cell losers not cleared by this mechanism were later eliminated from the basal cell layer by differentiation through the stratified cell layer. This is physiologically important because, when cell competition was disturbed, epithelial barrier function was compromised138. Whether this competitive cell behaviour is conserved throughout other epithelial tissues, including the airway lining, remains to be elucidated and warrants further investigation.
Transcriptomic technologies have enhanced our view of the cellular landscape in the human lung but there remain inherent limitations and many unanswered questions. Loss of fragile cell types in the process of sequencing can bias the analysis and variation in bioinformatic pipelines and cell annotation can hinder comparisons between different studies. Rare epithelial cell types have attracted much interest but, given their low frequency within the lung, the challenge will be to ascertain their contribution to diverse disease pathologies in the hope that we can harness and manipulate particular cells to promote lung health across the human life course.
References
Iwasaki, A., Foxman, E. F. & Molony, R. D. Early local immune defences in the respiratory tract. Nat. Rev. Immunol. 17, 7–20 (2017).
Rhodin, J. A. The ciliated cell. Ultrastructure and function of the human tracheal mucosa. Am. Rev. Respir. Dis. 93 (Suppl.), 1–15 (1966).
Breeze, R. G. & Wheeldon, E. B. The cells of the pulmonary airways. Am. Rev. Respir. Dis. 116, 705–777 (1977).
Widdicombe, J. H. Early studies on the surface epithelium of mammalian airways. Am. J. Physiol. Lung Cell Mol. Physiol. 317, L486–L495 (2019).
Mercer, R. R., Russell, M. L., Roggli, V. L. & Crapo, J. D. Cell number and distribution in human and rat airways. Am. J. Respir. Cell Mol. Biol. 10, 613–624 (1994).
Boers, J. E., Ambergen, A. W. & Thunnissen, F. B. Number and proliferation of basal and parabasal cells in normal human airway epithelium. Am. J. Respir. Crit. Care Med. 157, 2000–2006 (1998).
Boers, J. E., Ambergen, A. W. & Thunnissen, F. B. Number and proliferation of clara cells in normal human airway epithelium. Am. J. Respir. Crit. Care Med. 159, 1585–1591 (1999).
Huffnagle, G. B., Dickson, R. P. & Lukacs, N. W. The respiratory tract microbiome and lung inflammation: a two-way street. Mucosal Immunol. 10, 299–306 (2017).
Basil, M. C. et al. The cellular and physiological basis for lung repair and regeneration: past, present, and future. Cell Stem Cell 26, 482–502 (2020).
Vieira Braga, F. A. et al. A cellular census of human lungs identifies novel cell states in health and in asthma. Nat. Med. 25, 1153–1163 (2019). This was the first study to use scRNA-seq to analyse airway brushing and biopsy samples from human subjects.
Hie, B. et al. Computational methods for single-cell RNA sequencing. Annu. Rev. Biomed. Data Sci. 3, 339–364 (2020).
Mereu, E. et al. Benchmarking single-cell RNA-sequencing protocols for cell atlas projects. Nat. Biotechnol. 38, 747–755 (2020).
Luecken, M. D. & Theis, F. J. Current best practices in single-cell RNA-seq analysis: a tutorial. Mol. Syst. Biol. 15, e8746 (2019). This review provides a step-by-step discussion of the elements that comprise a typical scRNA-seq analysis workflow.
Schiller, H. B. et al. The human lung cell atlas: a high-resolution reference map of the human lung in health and disease. Am. J. Respir. Cell Mol. Biol. 61, 31–41 (2019).
Vento-Tormo, R. et al. Single-cell reconstruction of the early maternal-fetal interface in humans. Nature 563, 347–353 (2018).
Chua, R. L. et al. COVID-19 severity correlates with airway epithelium-immune cell interactions identified by single-cell analysis. Nat. Biotechnol. 38, 970–979 (2020).
Efremova, M., Vento-Tormo, M., Teichmann, S. A. & Vento-Tormo, R. CellPhoneDB: inferring cell-cell communication from combined expression of multi-subunit ligand-receptor complexes. Nat. Protoc. 15, 1484–1506 (2020).
Xu, Y. et al. Single-cell RNA sequencing identifies diverse roles of epithelial cells in idiopathic pulmonary fibrosis. JCI Insight 1, e90558 (2016).
Deprez, M. et al. A single-cell atlas of the human healthy airways. Am. J. Respir. Crit. Care Med. 202, 1636–1645 (2020). This study used scRNA-seq to profile airway epithelial cell types from brushings and biopsies taken at distinct locations throughout the nose, trachea and bronchi of healthy volunteers.
Madissoon, E. et al. scRNA-seq assessment of the human lung, spleen, and esophagus tissue stability after cold preservation. Genome Biol. 21, 1 (2019).
van den Brink, S. C. et al. Single-cell sequencing reveals dissociation-induced gene expression in tissue subpopulations. Nat. Methods 14, 935–936 (2017).
Plasschaert, L. W. et al. A single-cell atlas of the airway epithelium reveals the CFTR-rich pulmonary ionocyte. Nature 560, 377–381 (2018). This study used scRNA-seq to interrogate the gene expression profiles of mouse tracheal and cultured human bronchial epithelial cells, delineated the differentiation trajectories during homeostasis and tissue repair, and identified a novel cell type — the ‘ionocyte’.
Montoro, D. T. et al. A revised airway epithelial hierarchy includes CFTR-expressing ionocytes. Nature 560, 319–324 (2018). This seminal study utilized scRNA-seq to expose airway epithelial cell diversity and refine lineage hierarchies in the mouse trachea, notably discovering ‘hillock’ structures and the ‘ionocyte’.
Ruiz García, S. et al. Novel dynamics of human mucociliary differentiation revealed by single-cell RNA sequencing of nasal epithelial cultures. Development 146, dev177428 (2019).
Revinski, D. R. et al. CDC20B is required for deuterosome-mediated centriole production in multiciliated cells. Nat. Commun. 9, 4668 (2018).
Goldfarbmuren, K. C. et al. Dissecting the cellular specificity of smoking effects and reconstructing lineages in the human airway epithelium. Nat. Commun. 11, 2485 (2020). This study used scRNA-seq to evaluate the impact of cigarette smoking on each airway cell type in the human tracheal epithelium.
Adams, T. S. et al. Single-cell RNA-seq reveals ectopic and aberrant lung-resident cell populations in idiopathic pulmonary fibrosis. Sci. Adv. 6, eaba1983 (2020).
Habermann, A. C. et al. Single-cell RNA sequencing reveals profibrotic roles of distinct epithelial and mesenchymal lineages in pulmonary fibrosis. Sci. Adv. 6, eaba1972 (2020).
Treutlein, B. et al. Reconstructing lineage hierarchies of the distal lung epithelium using single-cell RNA-seq. Nature 509, 371–375 (2014).
Zaragosi, L. E., Deprez, M. & Barbry, P. Using single-cell RNA sequencing to unravel cell lineage relationships in the respiratory tract. Biochem. Soc. Trans. 48, 327–336 (2020).
Trapnell, C. et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 32, 381–386 (2014).
Saelens, W., Cannoodt, R., Todorov, H. & Saeys, Y. A comparison of single-cell trajectory inference methods. Nat. Biotechnol. 37, 547–554 (2019). This study evaluated the performance of 45 trajectory inference methods on different data sets and devised practical guidelines for their use.
La Manno, G. et al. RNA velocity of single cells. Nature 560, 494–498 (2018).
Bergen, V., Lange, M., Peidli, S., Wolf, F. A. & Theis, F. J. Generalizing RNA velocity to transient cell states through dynamical modeling. Nat. Biotechnol. 38, 1408–1414 (2020).
Haghverdi, L., Buettner, F. & Theis, F. J. Diffusion maps for high-dimensional single-cell analysis of differentiation data. Bioinformatics 31, 2989–2998 (2015).
Sungnak, W. et al. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat. Med. 26, 681–687 (2020).
Ziegler, C. G. K. et al. SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell 181, 1016–1035.e19 (2020).
Rock, J. R. et al. Basal cells as stem cells of the mouse trachea and human airway epithelium. Proc. Natl Acad. Sci. USA 106, 12771–12775 (2009).
Watson, J. K. et al. Clonal dynamics reveal two distinct populations of basal cells in slow-turnover airway epithelium. Cell Rep. 12, 90–101 (2015).
Schneider, C., O’Leary, C. E. & Locksley, R. M. Regulation of immune responses by tuft cells. Nat. Rev. Immunol. 19, 584–593 (2019).
Nadjsombati, M. S. et al. Detection of succinate by intestinal tuft cells triggers a type 2 innate immune circuit. Immunity 49, 33–41.e7 (2018).
von Moltke, J., Ji, M., Liang, H. E. & Locksley, R. M. Tuft-cell-derived IL-25 regulates an intestinal ILC2-epithelial response circuit. Nature 529, 221–225 (2016).
Tizzano, M. et al. Nasal chemosensory cells use bitter taste signaling to detect irritants and bacterial signals. Proc. Natl Acad. Sci. USA 107, 3210–3215 (2010). This article shows how chemosensory cells in the airways mediate reflex reactions to irritants, including bacterial quorum-sensing molecules, describing a mechanism by which bacterial population densities are limited by the local epithelial inflammatory response.
Ualiyeva, S. et al. Airway brush cells generate cysteinyl leukotrienes through the ATP sensor P2Y2. Sci. Immunol. 5, eaax7224 (2020).
Bankova, L. G. et al. The cysteinyl leukotriene 3 receptor regulates expansion of IL-25-producing airway brush cells leading to type 2 inflammation. Sci. Immunol. 3, eaat9453 (2018).
Krasteva, G. et al. Cholinergic chemosensory cells in the trachea regulate breathing. Proc. Natl Acad. Sci. USA 108, 9478–9483 (2011).
O’Leary, C. E., Schneider, C. & Locksley, R. M. Tuft cells-systemically dispersed sensory epithelia integrating immune and neural circuitry. Annu. Rev. Immunol. 37, 47–72 (2019).
Hollenhorst, M. I. et al. Tracheal brush cells release acetylcholine in response to bitter tastants for paracrine and autocrine signaling. FASEB J. 34, 316–332 (2020).
Kohanski, M. A. et al. Solitary chemosensory cells are a primary epithelial source of IL-25 in patients with chronic rhinosinusitis with nasal polyps. J. Allergy Clin. Immunol. 142, 460–469.e7 (2018).
Schneider, C. et al. A metabolite-triggered tuft cell-ILC2 circuit drives small intestinal remodeling. Cell 174, 271–284.e14 (2018).
Quigley, I. K., Stubbs, J. L. & Kintner, C. Specification of ion transport cells in the Xenopus larval skin. Development 138, 705–714 (2011).
Elborn, J. S. Cystic fibrosis. Lancet 388, 2519–2531 (2016).
Branchfield, K. et al. Pulmonary neuroendocrine cells function as airway sensors to control lung immune response. Science 351, 707–710 (2016). This article describes how PNECs act as rheostats to enable the lungs to sense and respond to changes in the inhaled environment.
Linnoila, R. I. Functional facets of the pulmonary neuroendocrine system. Lab. Invest. 86, 425–444 (2006).
Song, H. et al. Functional characterization of pulmonary neuroendocrine cells in lung development, injury, and tumorigenesis. Proc. Natl Acad. Sci. USA 109, 17531–17536 (2012).
Sui, P. et al. Pulmonary neuroendocrine cells amplify allergic asthma responses. Science 360, eaan8546 (2018).
Barrios, J. et al. Pulmonary neuroendocrine cells secrete γ-aminobutyric acid to induce goblet cell hyperplasia in primate models. Am. J. Respir. Cell Mol. Biol. 60, 687–694 (2019).
Barrios, J. et al. Early life allergen-induced mucus overproduction requires augmented neural stimulation of pulmonary neuroendocrine cell secretion. FASEB J. 31, 4117–4128 (2017).
Nagashima, H. et al. Neuropeptide CGRP limits group 2 innate lymphoid cell responses and constrains type 2 inflammation. Immunity 51, 682–695.e6 (2019).
Fahy, J. V. & Dickey, B. F. Airway mucus function and dysfunction. N. Engl. J. Med. 363, 2233–2247 (2010).
Johansen, F. E. & Kaetzel, C. S. Regulation of the polymeric immunoglobulin receptor and IgA transport: new advances in environmental factors that stimulate pIgR expression and its role in mucosal immunity. Mucosal Immunol. 4, 598–602 (2011).
Richmond, B. W. et al. Airway bacteria drive a progressive COPD-like phenotype in mice with polymeric immunoglobulin receptor deficiency. Nat. Commun. 7, 11240 (2016).
Lehmann, R. et al. Differential regulation of the transcriptomic and secretomic landscape of sensor and effector functions of human airway epithelial cells. Mucosal Immunol. 11, 627–642 (2018).
Brune, K., Frank, J., Schwingshackl, A., Finigan, J. & Sidhaye, V. K. Pulmonary epithelial barrier function: some new players and mechanisms. Am. J. Physiol. Lung Cell Mol. Physiol. 308, L731–L745 (2015).
Vermeer, P. D. et al. Segregation of receptor and ligand regulates activation of epithelial growth factor receptor. Nature 422, 322–326 (2003).
Fujino, N. et al. Sensing of apoptotic cells through Axl causes lung basal cell proliferation in inflammatory diseases. J. Exp. Med. 216, 2184–2201 (2019).
Ordovas-Montanes, J. et al. Allergic inflammatory memory in human respiratory epithelial progenitor cells. Nature 560, 649–654 (2018). This study demonstrates that basal stem cells in the human nasal epithelium acquire and retain allergic memory, contributing to chronic inflammatory disease.
White, S. R. Apoptosis and the airway epithelium. J. Allergy 2011, 948406 (2011).
Henson, P. M. & Hume, D. A. Apoptotic cell removal in development and tissue homeostasis. Trends Immunol. 27, 244–250 (2006).
Juncadella, I. J. et al. Apoptotic cell clearance by bronchial epithelial cells critically influences airway inflammation. Nature 493, 547–551 (2013).
Shaykhiev, R. & Crystal, R. G. Early events in the pathogenesis of chronic obstructive pulmonary disease. Smoking-induced reprogramming of airway epithelial basal progenitor cells. Ann. Am. Thorac. Soc. 11 (Suppl. 5), S252–S258 (2014).
Natoli, G. & Ostuni, R. Adaptation and memory in immune responses. Nat. Immunol. 20, 783–792 (2019).
Ordovas-Montanes, J., Beyaz, S., Rakoff-Nahoum, S. & Shalek, A. K. Distribution and storage of inflammatory memory in barrier tissues. Nat. Rev. Immunol. 20, 308–320 (2020).
Schleimer, R. P. Immunopathogenesis of chronic rhinosinusitis and nasal polyposis. Annu. Rev. Pathol. 12, 331–357 (2017).
Gibbs, J. E. et al. Circadian timing in the lung; a specific role for bronchiolar epithelial cells. Endocrinology 150, 268–276 (2009).
Gibbs, J. et al. An epithelial circadian clock controls pulmonary inflammation and glucocorticoid action. Nat. Med. 20, 919–926 (2014).
Man, K., Loudon, A. & Chawla, A. Immunity around the clock. Science 354, 999–1003 (2016).
Gibbs, J. E. et al. The nuclear receptor REV-ERBα mediates circadian regulation of innate immunity through selective regulation of inflammatory cytokines. Proc. Natl Acad. Sci. USA 109, 582–587 (2012).
Pariollaud, M. et al. Circadian clock component REV-ERBalpha controls homeostatic regulation of pulmonary inflammation. J. Clin. Invest. 128, 2281–2296 (2018).
Schupp, J. C. et al. Single cell transcriptional archetypes of airway inflammation in cystic fibrosis. Am. J. Respir. Crit. Care Med. 202, 1419–1429 (2020).
Morse, C. et al. Proliferating SPP1/MERTK-expressing macrophages in idiopathic pulmonary fibrosis. Eur. Respir. J. 54, 1802441 (2019).
Liao, M. et al. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat. Med. 26, 842–844 (2020).
Raredon, M. S. B. et al. Single-cell connectomic analysis of adult mammalian lungs. Sci. Adv. 5, eaaw3851 (2019).
Yan, X. et al. Non-invasive analysis of the sputum transcriptome discriminates clinical phenotypes of asthma. Am. J. Respir. Crit. Care Med. 191, 1116–1125 (2015).
Yu, Y. R. et al. Flow cytometric analysis of myeloid cells in human blood, bronchoalveolar lavage, and lung tissues. Am. J. Respir. Cell Mol. Biol. 54, 13–24 (2016).
Guilliams, M. et al. Alveolar macrophages develop from fetal monocytes that differentiate into long-lived cells in the first week of life via GM-CSF. J. Exp. Med. 210, 1977–1992 (2013).
Hashimoto, D. et al. Tissue-resident macrophages self-maintain locally throughout adult life with minimal contribution from circulating monocytes. Immunity 38, 792–804 (2013).
Byrne, A. J. et al. Dynamics of human monocytes and airway macrophages during healthy aging and after transplant. J. Exp. Med. 217, e20191236 (2020).
Puttur, F., Gregory, L. G. & Lloyd, C. M. Airway macrophages as the guardians of tissue repair in the lung. Immunol. Cell Biol. 97, 246–257 (2019).
Westphalen, K. et al. Sessile alveolar macrophages communicate with alveolar epithelium to modulate immunity. Nature 506, 503–506 (2014).
Moon, H. G. et al. Airway epithelial cell-derived colony stimulating factor-1 promotes allergen sensitization. Immunity 49, 275–287.e5 (2018).
Masopust, D. & Soerens, A. G. Tissue-resident T cells and other resident leukocytes. Annu. Rev. Immunol. 37, 521–546 (2019).
Turner, D. L. et al. Lung niches for the generation and maintenance of tissue-resident memory T cells. Mucosal Immunol. 7, 501–510 (2014).
Oja, A. E. et al. Trigger-happy resident memory CD4+ T cells inhabit the human lungs. Mucosal Immunol. 11, 654–667 (2018).
Kumar, B. V. et al. Human tissue-resident memory T cells are defined by core transcriptional and functional signatures in lymphoid and mucosal sites. Cell Rep. 20, 2921–2934 (2017).
Takamura, S. et al. Interstitial-resident memory CD8+ T cells sustain frontline epithelial memory in the lung. J. Exp. Med. 216, 2736–2747 (2019).
Snyder, M. E. et al. Generation and persistence of human tissue-resident memory T cells in lung transplantation. Sci. Immunol. 4, eaav5581 (2019). This study demonstrates that donor TRM cells persisted around the airways of lung transplant recipients for over 1 year.
Higgins, J. M. G. et al. Direct and regulated interaction of integrin αEβ7 with E-cadherin. J. Cell Biol. 140, 197–210 (1998).
Shenoy, A. T. et al. Lung CD4+ resident memory T cells remodel epithelial responses to accelerate neutrophil recruitment during pneumonia. Mucosal Immunol. 13, 334–343 (2020).
Turner, D. L. et al. Biased generation and in situ activation of lung tissue-resident memory CD4 T cells in the pathogenesis of allergic asthma. J. Immunol. 200, 1561–1569 (2018).
Hayward, S. L. et al. Environmental cues regulate epigenetic reprogramming of airway-resident memory CD8+ T cells. Nat. Immunol. 21, 309–320 (2020).
Hinks, T. S. et al. Steroid-induced deficiency of mucosal-associated invariant T cells in the chronic obstructive pulmonary disease lung. Implications for nontypeable haemophilus influenzae infection. Am. J. Respir. Crit. Care Med. 194, 1208–1218 (2016).
Toubal, A., Nel, I., Lotersztajn, S. & Lehuen, A. Mucosal-associated invariant T cells and disease. Nat. Rev. Immunol. 19, 643–657 (2019).
Wang, H. et al. MAIT cells protect against pulmonary Legionella longbeachae infection. Nat. Commun. 9, 3350 (2018).
Hinks, T. S. C. et al. Activation and in vivo evolution of the MAIT cell transcriptome in mice and humans reveals tissue repair functionality. Cell Rep. 28, 3249–3262.e5 (2019).
Broggi, A. et al. Type III interferons disrupt the lung epithelial barrier upon viral recognition. Science 369, 706–712 (2020).
Major, J. et al. Type I and III interferons disrupt lung epithelial repair during recovery from viral infection. Science 369, 712–717 (2020).
Channappanavar, R. et al. Dysregulated type I interferon and inflammatory monocyte-macrophage responses cause lethal pneumonia in SARS-CoV-infected mice. Cell Host Microbe 19, 181–193 (2016).
Blanco-Melo, D. et al. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell 181, 1036–1045.e9 (2020).
Zhang, Q. et al. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science 370, eabd4570 (2020).
Bastard, P. et al. Auto-antibodies against type I IFNs in patients with life-threatening COVID-19. Science 370, eabd4585 (2020).
Prescott, S. L., Umans, B. D., Williams, E. K., Brust, R. D. & Liberles, S. D. An airway protection program revealed by sweeping genetic control of vagal afferents. Cell 181, 574–589.e14 (2020).
Ordovas-Montanes, J. et al. The regulation of immunological processes by peripheral neurons in homeostasis and disease. Trends Immunol. 36, 578–604 (2015).
Godinho-Silva, C., Cardoso, F. & Veiga-Fernandes, H. Neuro-immune cell units: a new paradigm in physiology. Annu. Rev. Immunol. 37, 19–46 (2019).
Solis, A. G. et al. Mechanosensation of cyclical force by PIEZO1 is essential for innate immunity. Nature 573, 69–74 (2019). This article describes how mechanical forces influence innate immune pathways in the lungs via the expression of PIEZO1, a mechanically activated ion channel.
Coleridge, H. M., Coleridge, J. C. & Schultz, H. D. Afferent pathways involved in reflex regulation of airway smooth muscle. Pharmacol. Ther. 42, 1–63 (1989).
Slebos, D. J. et al. Safety and adverse events after targeted lung denervation for symptomatic moderate to severe chronic obstructive pulmonary disease (AIRFLOW). A multicenter randomized controlled clinical trial. Am. J. Respir. Crit. Care Med. 200, 1477–1486 (2019).
Talbot, S. et al. Silencing nociceptor neurons reduces allergic airway inflammation. Neuron 87, 341–354 (2015).
Trankner, D., Hahne, N., Sugino, K., Hoon, M. A. & Zuker, C. Population of sensory neurons essential for asthmatic hyperreactivity of inflamed airways. Proc. Natl Acad. Sci. USA 111, 11515–11520 (2014).
Kistemaker, L. E., Slebos, D. J., Meurs, H., Kerstjens, H. A. & Gosens, R. Anti-inflammatory effects of targeted lung denervation in patients with COPD. Eur. Respir. J. 46, 1489–1492 (2015).
Alpizar, Y. A. et al. TRPV4 activation triggers protective responses to bacterial lipopolysaccharides in airway epithelial cells. Nat. Commun. 8, 1059 (2017).
Baral, P. et al. Nociceptor sensory neurons suppress neutrophil and γδ T cell responses in bacterial lung infections and lethal pneumonia. Nat. Med. 24, 417–426 (2018). This study shows how lung sensory neurons contribute to innate host defence mechanisms against lethal bacterial infections.
Klose, C. S. & Artis, D. Neuronal regulation of innate lymphoid cells. Curr. Opin. Immunol. 56, 94–99 (2019).
Huh, J. R. & Veiga-Fernandes, H. Neuroimmune circuits in inter-organ communication. Nat. Rev. Immunol. 20, 217–228 (2020).
Germain, R. N. & Huang, Y. ILC2s - resident lymphocytes pre-adapted to a specific tissue or migratory effectors that adapt to where they move? Curr. Opin. Immunol. 56, 76–81 (2019).
Wallrapp, A. et al. The neuropeptide NMU amplifies ILC2-driven allergic lung inflammation. Nature 549, 351–356 (2017).
Klose, C. S. N. et al. The neuropeptide neuromedin U stimulates innate lymphoid cells and type 2 inflammation. Nature 549, 282–286 (2017).
Prendergast, C. E., Morton, M. F., Figueroa, K. W., Wu, X. & Shankley, N. P. Species-dependent smooth muscle contraction to Neuromedin U and determination of the receptor subtypes mediating contraction using NMU1 receptor knockout mice. Br. J. Pharmacol. 147, 886–896 (2006).
Puttur, F. et al. Pulmonary environmental cues drive group 2 innate lymphoid cell dynamics in mice and humans. Sci. Immunol. 4, eaav7638 (2019). This study shows that extracellular matrix influences cell dynamics in vivo and in vitro, changing the course of inflammation.
Chen, H. et al. Mechanosensing by the alpha6-integrin confers an invasive fibroblast phenotype and mediates lung fibrosis. Nat. Commun. 7, 12564 (2016).
Wu, H. et al. Progressive pulmonary fibrosis is caused by elevated mechanical tension on alveolar stem cells. Cell 180, 107–121.e17 (2020).
Duclos, G. E. et al. Characterizing smoking-induced transcriptional heterogeneity in the human bronchial epithelium at single-cell resolution. Sci. Adv. 5, eaaw3413 (2019).
Rodriques, S. G. et al. Slide-seq: a scalable technology for measuring genome-wide expression at high spatial resolution. Science 363, 1463–1467 (2019).
Angelidis, I. et al. An atlas of the aging lung mapped by single cell transcriptomics and deep tissue proteomics. Nat. Commun. 10, 963 (2019).
Hiemstra, P. S., Tetley, T. D. & Janes, S. M. Airway and alveolar epithelial cells in culture. Eur. Respir. J. 54, 1900742 (2019).
Benam, K. H. et al. Small airway-on-a-chip enables analysis of human lung inflammation and drug responses in vitro. Nat. Methods 13, 151–157 (2016).
Pardo-Saganta, A. et al. Injury induces direct lineage segregation of functionally distinct airway basal stem/progenitor cell subpopulations. Cell Stem Cell 16, 184–197 (2015).
Ellis, S. J. et al. Distinct modes of cell competition shape mammalian tissue morphogenesis. Nature 569, 497–502 (2019).
Ramachandran, P. et al. Resolving the fibrotic niche of human liver cirrhosis at single-cell level. Nature 575, 512–518 (2019).
Nakajima, M. et al. Immunohistochemical and ultrastructural studies of basal cells, Clara cells and bronchiolar cuboidal cells in normal human airways. Pathol. Int. 48, 944–953 (1998).
Pack, R. J., Al-Ugaily, L. H., Morris, G. & Widdicombe, J. G. The distribution and structure of cells in the tracheal epithelium of the mouse. Cell Tissue Res. 208, 65–84 (1980).
Liu, X. & Engelhardt, J. F. The glandular stem/progenitor cell niche in airway development and repair. Proc. Am. Thorac. Soc. 5, 682–688 (2008).
Tata, A. et al. Myoepithelial cells of submucosal glands can function as reserve stem cells to regenerate airways after injury. Cell Stem Cell 22, 668–683.e6 (2018).
Rock, J. R., Randell, S. H. & Hogan, B. L. Airway basal stem cells: a perspective on their roles in epithelial homeostasis and remodeling. Dis. Model. Mech. 3, 545–556 (2010).
Tao, L. & Reese, T. A. Making mouse models that reflect human immune responses. Trends Immunol. 38, 181–193 (2017).
Beura, L. K. et al. Normalizing the environment recapitulates adult human immune traits in laboratory mice. Nature 532, 512–516 (2016).
Abolins, S. et al. The comparative immunology of wild and laboratory mice, Mus musculus domesticus. Nat. Commun. 8, 14811 (2017).
Acknowledgements
CML is a Wellcome Senior Fellow in Basic Biomedical Sciences (107059/z/15/z), RJH is funded through the Imperial College Clinician-Investigator Scholarship programme.
Author information
Authors and Affiliations
Contributions
The authors contributed equally to all aspects of the article.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information
Nature Reviews Immunology thanks J. Kolls and J. Ordovas-Montanes for their contribution to the peer review of this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Glossary
- Bronchoscopy
-
A procedure in which a fibre-optic camera is used to visualize the airways.
- Transcriptome
-
All the RNA transcripts expressed in a cell or population of cells.
- Human Cell Atlas Consortium
-
An international community of scientists collaborating with the primary goal of characterizing all human cell types by unique gene expression profile, developmental trajectories and spatial localization.
- Idiopathic pulmonary fibrosis
-
(IPF). Progressive, scarring lung disease driven by alveolar epithelial cell injury, fibroblast activation and excessive extracellular matrix deposition in the interstitium, impairing gas exchange.
- Bronchial brushings
-
A procedure in which a brush is used through a bronchoscope to obtain cells lining the airway under direct visualization.
- Endobronchial biopsy
-
A procedure in which a small biopsy is performed using forceps through a bronchoscope to obtain a sample of airway wall cells under direct visualization.
- Air–liquid interface cultures
-
Epithelial cells are cultured on a microporous membrane in a Transwell plate and exposed to air to allow mucociliary differentiation.
- Asthma
-
Lung disease characterized by chronic airway inflammation and remodeling, causing variable expiratory airflow obstruction and airway hyperresponsiveness to a range of stimuli. Often associated with allergen sensitization.
- Turbinates
-
Small structures inside the nose that filter, warm and humidify air as it passes through the nostrils into the lungs.
- Calcitonin gene-related peptide
-
(CGRP). A neuropeptide released from sensory nerve fibres.
- GABA
-
γ-aminobutyric acid, an inhibitory neurotransmitter.
- Pattern recognition receptors
-
Specialized immune receptors that recognize conserved molecular structures on bacteria and viruses. These include Toll-like receptors.
- Bronchoalveolar lavage
-
(BAL). A procedure carried out during bronchoscopy in which sterile normal saline is instilled into a subsegment of the lung and collected for analysis. It provides information about the immune cell repertoire in the alveolar compartment of the lung.
- Ccsp-Cre mouse strain
-
Cre recombinase expression under the club cell secretory protein (Ccsp) gene promoter to enable airway cell-specific gene ablation.
- Chronic obstructive pulmonary disease
-
(COPD). Lung disease characterized by chronic airflow obstruction due to inflammation and remodelling of the small airways and destruction of the lung parenchyma.
- Chronic rhinosinusitis
-
(CRS). Type 2-mediated mucosal inflammation of the nose and paranasal sinuses.
- ATAC-seq
-
A next-generation sequencing technique allowing the analysis of open or accessible chromatin regions of the genome.
- Parabiosis
-
Surgical procedure in which two animals are joined so that they share the same circulation.
- Transient receptor potential vanilloid type 1
-
(TRPV1). Non-selective cation channel activated by diverse stimuli, including capsaicin, heat and low pH, and expressed on a variety of cell types.
- Neuromedin U
-
(NMU). A neuropeptide with a diverse range of functions, including smooth muscle contraction, pain perception, regulation of blood pressure, stress responses and feeding behaviour.
- Vasoactive intestinal peptide
-
(VIP). A neuropeptide.
Rights and permissions
About this article
Cite this article
Hewitt, R.J., Lloyd, C.M. Regulation of immune responses by the airway epithelial cell landscape. Nat Rev Immunol 21, 347–362 (2021). https://doi.org/10.1038/s41577-020-00477-9
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41577-020-00477-9
This article is cited by
-
Bufei Yishen formula protects the airway epithelial barrier and ameliorates COPD by enhancing autophagy through the Sirt1/AMPK/Foxo3 signaling pathway
Chinese Medicine (2024)
-
Biochemical and transcriptomic evaluation of a 3D lung organoid platform for pre-clinical testing of active substances targeting senescence
Respiratory Research (2024)
-
Single-cell division tracing and transcriptomics reveal cell types and differentiation paths in the regenerating lung
Nature Communications (2024)
-
Lung microbiome: new insights into the pathogenesis of respiratory diseases
Signal Transduction and Targeted Therapy (2024)
-
Enabling methanol fixation of pediatric nasal wash during respiratory illness for single cell sequencing in comparison with fresh samples
Pediatric Research (2024)